X-Chromosome Inactivation (XCI) is one of the best-characterized epigenetic phenomena where long noncoding RNAs are key players that regulate gene expression. Female mammals (XX) have two X-chromosomes, and one X is randomly chosen for transcriptional silencing in order to equalize the expression of X-linked genes compared to males (XY). Thus females are mosaic for X-chromosome expression, where a cell will express the maternal or paternal X. One great example of female X-mosaicism are calico cats.
best-characterized epigenetic phenomena where long noncoding RNAs are key players that regulate gene expression. Female mammals (XX) have two X-chromosomes, and one X is randomly chosen for transcriptional silencing in order to equalize the expression of X-linked genes compared to males (XY). Thus females are mosaic for X-chromosome expression, where a cell will express the maternal or paternal X. One great example of female X-mosaicism are calico cats.
The X-chromosome contains about 1000 genes with roles in development, reproduction, metabolism, and immunity. Mammalian females (including humans) are healthier and have longer life-spans than males. Females have better survival rates than males from illnesses caused by infectious diseases, injury, or sepsis. The X-chromosome plays an important role in the hyper-responsive female immune system because this chromosome has the highest density of immune-related genes. Females are also more susceptible to a number of autoimmune diseases, suggesting that the enhanced immune function may be related to the loss of self-tolerance.
The X-chromosome contains about 1000 genes with roles in development, reproduction, metabolism, and immunity. Mammalian females (including humans) are healthier and have longer life-spans than males. Females have better survival rates than males from illnesses caused by infectious diseases, injury, or sepsis. The X-chromosome plays an important role in the hyper-responsive female immune system because this chromosome has the highest density of immune-related genes. Females are also more susceptible to a number of autoimmune diseases, suggesting that the enhanced immune function may be related to the loss of self-tolerance.
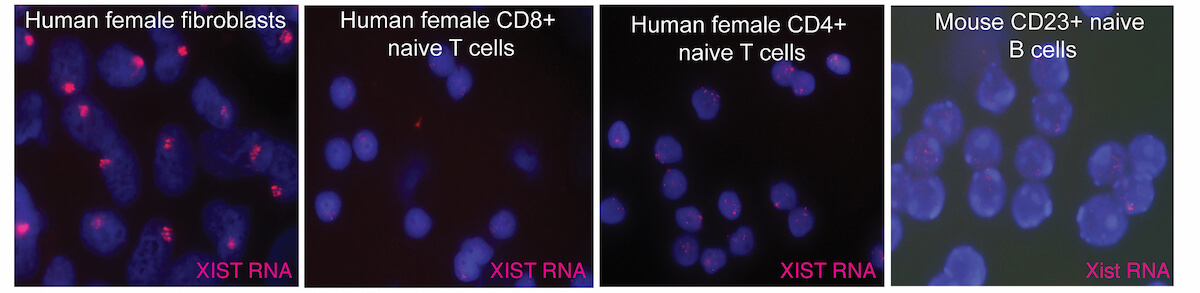
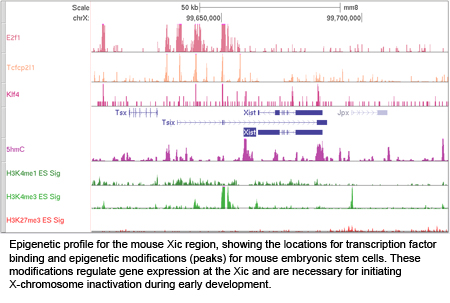
XCI occurs during early female embryonic development, and is initiated by expression of Xist RNA which recruits heterochromatic complexes for transcriptional silencing. Xist RNA and heterochromatin modifications associate with the inactive X chromosome with each cell division, and can be visualized using fluorescence microscopy. Xist RNA also functions to maintain XCI in somatic cells.
We recently discovered that XCI is maintained differently in female lymphocytes. Naïve T and B cells, which are quiescent, lack Xist RNA and heterochromatin marks on the inactive X. After lymphocyte stimulation, these epigenetic modifications re-localize back to the inactive X, as the cells re-enter the cell cycle and actively proliferate. This dynamic mechanism for XCI maintenance is unique to immune cells, and our lab investigates the molecular details of this process. The major areas of investigation in our lab are: (1) to understand how XCI is maintained in female lymphocytes, and how this contributes to female-biased autoimmunity; (2) to investigate epigenetic mechanisms involving long noncoding RNAs during early human development and placental progenitors.
Research Techniques
- Microscopy: Nuclear imaging of Xist RNA and the X-chromosome using RNA and DNA in situ hybridization (FISH); immunofluorescence detection of heterochromatic histone tail modifications
- Allele-specific transcriptional & epigenomic profiling of the inactive X and active X chromosome across somatic cell types (RNA-seq, CUT&RUN, ChIP-seq, Hi-C, DNA methylation)
- Mouse models of autoimmune disease, where animals spontaneously develop lupus-like disease symptoms. Induction of autoimmune disease features using pristane or imiquimod
- Human immune cell profiling of patients with female-biased autoimmune diseases, investigating epigenomic features of the X chromosomes and sex-biased differences with immune cell distribution & cell function
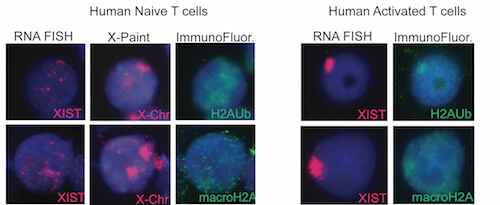
XCI maintenance is different in mammalian female lymphocytes
We have discovered that the inactive X chromosome lacks the typical heterochromatic modifications in female mature naïve T and B cells. This is the first example of physiologically-relevant female somatic cells that don’t have XIST/Xist RNA, H3K27me3, macroH2A, H2A-ubiquitin, and H4K20me modifications enriched on the inactive X. We also observed that in vitro activation of T and B cells stimulates the return of XIST/Xist RNA and some heterochromatic modifications back to the inactive X.
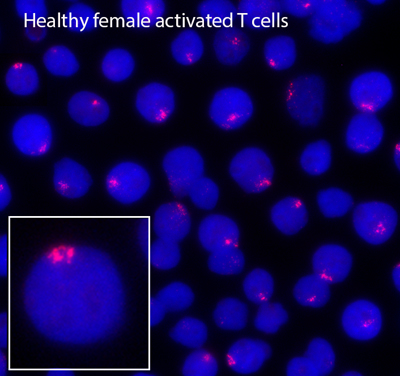
Using loss of function assays, we identified protein factors responsible for localizing XIST/Xist RNA back to the inactive X during lymphocyte activation. YY1 and hnRNP U, known XIST RNA binding proteins, return the XIST/Xist transcripts back to the inactive X. Elucidating the molecular mechanism of XIST/Xist RNA relocalization back to the inactive X, and how this transcript initiates the return of heterochromatin modifications, will reveal how the memory of transcriptional silencing is maintained after cell division in lymphocytes.
XIST/Xist RNA localization on the inactive X is perturbed in lymphocytes from female lupus patients and lupus mouse models
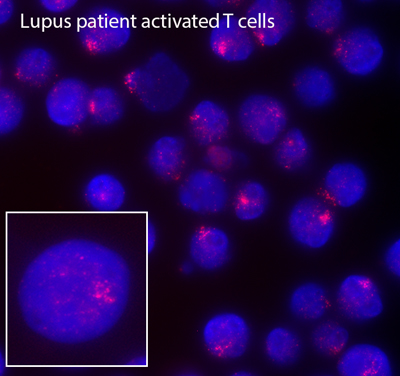
Remarkably, we have recently found that female lupus patients exhibit differences with XCI compared to healthy controls, which may explain the female bias of autoimmune disorders and why female lupus patients express higher levels of X-linked autoimmunity related genes compared to male patients. SLE-patient B cells have fewer typical XIST RNA clouds and more cells with dispersed patterns of XIST RNA or completely lacking XIST localization. Abnormal Xist RNA localization suggests partial reactivation of the inactive X-chromosome, which could increase gene expression. Using RNA FISH, we demonstrated that female lupus T and B cells frequently exhibit biallelic expression of CD40LG, CXCR3 and TLR7, which correlates with elevated expression of these genes. Moving forward, we are investigating causality of partial X-reactivation for lupus pathogenesis using mouse models. These will be the first experiments that directly connect transcription from the inactive X to lupus pathology.
 |
 |
Diversity of XCI maintenance mechanisms in immune cells
Our lab recently found that other immune cell populations, including dendritic cells, NK cells, and monocytes, have different mechanisms to maintain XCI. Using single-cell microscopy to image Xist RNA and heterochromatin modifications, we observed that monocytes and NK cells have faint Xist RNA ‘clouds’ and H3K27me3 foci, yet plasmacytoid dentritic cells (p-DCs) completely lack Xist RNA signals and enrichment of this histone mark. Unlike in humans, mouse p-DCs do not exhibit sex differences with interferon alpha production, and interferon signature gene expression in p-DCs is similar between males and females. Thus, immune cells use diverse (& unknown) mechanisms to maintain XCI, and we propose that these mechanisms are susceptible to error, thereby contributing to sex-linked autoimmune diseases. Our lab is actively investigating the molecular mechanisms that maintain XCI in a variety of immune cells.
Jiwrajka N, Toothacre NE, Beethem ZT, Sting S, Forsyth KS, Dubin AH, Driscoll A, Stohl W, Anguera MC. (2023) “Impaired dynamic X-chromosome inactivation maintenance in T cells is a feature of spontaneous murine SLE that is exacerbated in female-biased models.” J. Autoimmun. Sept; 139:103084. PMID: 37399593
Sierra I, Pyfrom S, Weiner A, Zhao G, Driscoll A, Yu X, Gregory BD, Vaughan AE, Anguera MC. (2022) “Unusual X chromosome inactivation maintenance in female alveolar type 2 cells is correlated with increased numbers of X-linked escape genes and sex-biased gene expression”. Stem Cell Reports. Dec 27:S2213-6711(22)00594-X. PMID: 36638790
Lovell, C. D. and Anguera, M.C. (2022) “Long noncoding RNAs that function in nutrition: Lnc-ing nutritional cues to metabolic pathways.” Annu Rev Nutr. 2022 Apr 18. PMID: 35436418
Jiwrajka, N. and Anguera M.C. (2022) “The X in seX-biased immunity and autoimmune rheumatic disease”. J Exp Med. Jun 6; 291(6):e20211487. PMID:35510951
Volkmann ER, Siegfried J, Lahm T, Ventetulo C, Mathai SC, Stenn V, Herzog E, Shansky R, Anguera MC, Danoff S, Giles JT, Lee YC, Drake W, Maier LA, Lachowicz-Scroggins M, Park H, Banerjee K, Fessel J, Reineck L, Vuga L, Crouser E, Feghali-Bostwick C. (2022) “NHLBI Report: Impact of Sex and Gender on autoimmune lung disease, opportunities for future research”. Am J Respir Crit Care Med. May 13. PMID: 35549658
Pyfrom S, Paneru B, Knoxx, J, Posso S, Buckner J, Cancro M, Anguera MC. (2021) “The dynamic epigenetic regulation of the inactive X chromosome in healthy human B cells is dysregulated in lupus patients”. Proc Natl Acad Sci. Jun 15;118 (24). PMID: 34103397
Forsyth, Katherine S. and Anguera, M.C. (2021) “Time to get ill: the intersection of viral infections, sex, and the X chromosome.” Curr Opin Physiol. Feb; 19:62-72. PMCID: PMC7553007
Chang S, Hur SK, Naveh NSS, Thorvaldsen JL, French DL, Gagne AL, Jobaliya CD, Anguera MC, Bartolomei MS, Kalish JM. (2021) “Derivation and investigation of the first human cell-based model of Beckwith-Wiedemann syndrome”. Epigenetics. Dec 16(12):1295-1305. PMID: 33300436
Syrett CM, Sierra I, Beethem ZT, Dubin AH, and Anguera M.C. (2019) “Loss of epigenetic modifications on the inactive X chromosome and sex-biased gene expression profiles in B cells from NZB/W F1 mice with lupus-like disease.” J Autoimmun. Nov 25:102357. PMID: 31780316
Syrett Camille M, Anguera Montserrat C:
When the balance is broken: X-linked gene dosage from two X chromosomes and female-biased autoimmunity. [PMID 31125996] Journal of leukocyte biology May 2019.
Kotzin Jonathan J, Iseka Fany, Wright Jasmine, Basavappa Megha G, Clark Megan L, Ali Mohammed-Alkhatim, Abdel-Hakeem Mohamed S, Robertson Tanner F, Mowel Walter K, Joannas Leonel, Neal Vanessa D, Spencer Sean P, Syrett Camille M, Anguera Montserrat C, Williams Adam, Wherry E John, Henao-Mejia Jorge: The long noncoding RNA regulates CD8 T cells in response to viral infection.[PMID 31138702] Proceedings of the National Academy of Sciences of the United States of America 116(24): 11916-11925, Jun 2019.
Syrett Camille M, Paneru Bam, Sandoval-Heglund Donavon, Wang Jianle, Banerjee Sarmistha, Sindhava Vishal, Behrens Edward M, Atchison Michael, Anguera Montserrat C: Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases. [PMID 30944248 JCI insight 4(7), Apr 2019.
Syrett Camille M, Sindhava Vishal, Sierra Isabel, Dubin Aimee H, Atchison Michael, Anguera Montserrat C: Diversity of Epigenetic Features of the Inactive X-Chromosome in NK Cells, Dendritic Cells, and Macrophages. [PMID 30671059] Frontiers in immunology 9: 3087, 2018.
Le Coz Carole, Trofa Melissa, Syrett Camille M, Martin Anna, Jyonouchi Harumi, Jyonouchi Soma, Anguera Montserrat C, Romberg Neil: CD40LG duplication-associated autoimmune disease is silenced by nonrandom X-chromosome inactivation.[PMID 29499223] The Journal of allergy and clinical immunology 141(6): 2308-2311.e7, Jun 2018.
Syrett Camille M, Sierra Isabel, Berry Corbett L, Beiting Daniel, Anguera Montserrat C: Sex-Specific Gene Expression Differences Are Evident in Human Embryonic Stem Cells and During In Vitro Differentiation of Human Placental Progenitor Cells. [PMID 29993333] Stem cells and development 27(19): 1360-1375, Oct 2018.
Wang Jianle, Anguera Montserrat C: In Vitro Differentiation of Human Pluripotent Stem Cells into Trophoblastic Cells. [PMID 28362386] Journal of visualized experiments : JoVE(121), Mar 2017.
Syrett Camille M, Sindhava Vishal, Hodawadekar Suchita, Myles Arpita, Liang Guanxiang, Zhang Yue, Nandi Satabdi, Cancro Michael, Atchison Michael, Anguera Montserrat C: Loss of Xist RNA from the inactive X during B cell development is restored in a dynamic YY1-dependent two-step process in activated B cells. [PMID 28991910] PLoS genetics 13(10): e1007050, Oct 2017.
Penkala Ian, Wang Jianle, Syrett Camille M, Goetzl Laura, López Carolina B, Anguera Montserrat C: lncRHOXF1, a Long Noncoding RNA from the X Chromosome That Suppresses Viral Response Genes during Development of the Early Human Placenta. [PMID 27066803] Molecular and cellular biology 36(12): 1764-75, Jun 2016.
Wang Jianle, Syrett Camille M, Kramer Marianne C, Basu Arindam, Atchison Michael L, Anguera Montserrat C: Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. [PMID 27001848] Proceedings of the National Academy of Sciences of the United States of America 113(14): E2029-38, Apr 2016.
Luo Mengcheng, Zhou Jian, Leu N Adrian, Abreu Carla M, Wang Jianle, Anguera Montserrat C, de Rooij Dirk G, Jasin Maria, Wang P Jeremy: Polycomb protein SCML2 associates with USP7 and counteracts histone H2A ubiquitination in the XY chromatin during male meiosis. [PMID 25634095] PLoS genetics 11(1): e1004954, Jan 2015.
Lessing D, Anguera MC, Lee JT.: X chromosome inactivation and epigenetic responses to cellular reprogramming. [PMID 23662665] Annu Rev Genomics Hum Genet. 14: 85-110, May 2013.
Anguera Montserrat C, Sadreyev Ruslan, Zhang Zhaoqing, Szanto Attila, Payer Bernhard, Sheridan Steven D, Kwok Showming, Haggarty Stephen J, Sur Mriganka, Alvarez Jason, Gimelbrant Alexander, Mitalipova Maisam, Kirby James E, Lee Jeannie T: Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. [PMID 22770242] Cell stem cell 11(1): 75-90, Jul 2012.
Anguera Montserrat C, Ma Weiyuan, Clift Danielle, Namekawa Satoshi, Kelleher Raymond J, Lee Jeannie T: Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. [PMID 21912526] PLoS genetics 7(9): e1002248, Sep 2011.
Kim Daniel H, Jeon Yesu, Anguera Montserrat C, Lee Jeannie T: X-chromosome epigenetic reprogramming in pluripotent stem cells via noncoding genes. [PMID 21376830] Seminars in cell & developmental biology 22(4): 336-42, Jun 2011.
Field Martha S, Anguera Montserrat C, Page Rodney, Stover Patrick J: 5,10-Methenyltetrahydrofolate synthetase activity is increased in tumors and modifies the efficacy of antipurine LY309887. [PMID 19022216] Archives of biochemistry and biophysics 481(2): 145-50, Jan 2009.
Anguera Montserrat C, Liu Matthew, Avruch Joseph, Lee Jeannie T: Characterization of two Mst1-deficient mouse models. [PMID 18942145] Developmental dynamics : an official publication of the American Association of Anatomists 237(11): 3424-34, Nov 2008.