New Penn Vet Studies Made Possible by Robert and Hope Sheft
Through Penn Vet’s Center for Host-Microbial Interactions, researchers are exploring the microbiome of animals in order to benefit both animal and human health. Three new studies will explore how microbes impact alcoholic liver disease, infections caused by Salmonella, and periodontal disease. These projects are funded by a generous gift from Robert and Hope Sheft.
“Through the generosity of the Shefts, we are able to support exciting new areas of research that have broad implications for both animal and human health,” said Dr. Christopher Hunter, Director of the Center for Host-Microbial Interactions.
The role of the intestinal microbiome in alcoholic liver disease
Dr. Narayan Avadhani, the Harriet Ellison Woodward Professor of Biochemistry at Penn Vet, will examine the role of the intestinal microbiome and intestinal permeability in alcoholic liver disease.
 Alcohol consumption has been implicated in a range of human diseases, including alcoholic liver disease, liver cancer, and pancreatitis. Current alcohol toxicity research focuses on the enzymes that break down alcohol. However, this strategy has not been successful in understanding the mechanism of alcoholic liver disease or its treatment.
Alcohol consumption has been implicated in a range of human diseases, including alcoholic liver disease, liver cancer, and pancreatitis. Current alcohol toxicity research focuses on the enzymes that break down alcohol. However, this strategy has not been successful in understanding the mechanism of alcoholic liver disease or its treatment.
Avadhani hypothesizes that gut bacteria play a major role in causing disease by making the gut more permeable. When excess alcohol is consumed, the composition of the gut bacteria changes, compromising intestinal barrier function and allowing toxins to enter the bloodstream and cause disease.
Avadhani will study the relationship between changes in the gut communities of bacteria and gut barrier function. His research could lead to successful treatments in humans whose gut bacteria have been altered from excess alcohol consumption. By replacing the bacteria they are missing, the gut may be repaired.
The molecular mechanisms that lead to serious infections caused by Salmonella
Dr. Dieter Schifferli, Professor of Microbiology at Penn Vet, will sequence the Salmonella genome in order to determine the molecular mechanisms that cause serious septic infections in humans and livestock.
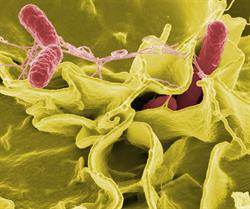 Salmonella remains the most frequent bacterial cause of foodborne disease in the U.S. and of typhoid fever in developing countries. In certain strains of Salmonella, bacteria remain in the intestine, causing mild gastrointestinal illness. In other strains, bacteria migrate from the intestine and enter the bloodstream, causing serious septic infections that can result in morbidity or even death in both livestock and humans.
Salmonella remains the most frequent bacterial cause of foodborne disease in the U.S. and of typhoid fever in developing countries. In certain strains of Salmonella, bacteria remain in the intestine, causing mild gastrointestinal illness. In other strains, bacteria migrate from the intestine and enter the bloodstream, causing serious septic infections that can result in morbidity or even death in both livestock and humans.
The molecular mechanisms that allow bacteria to leave a host’s intestines and cause systemic infection are poorly understood. Using various strains collected from agricultural cases, Schifferli will sequence the genomes of systemic Salmonella strains to discover the genetic factors that arm these bacteria to migrate out of the gut. He aims to discover whole pieces of the genome that are important in determining how bacteria become more or less virulent. This information could support the development of new drugs or vaccines against septicemic Salmonella.
The role of periodontal disease in the oral microbiome
Dr. Tom Schaer, Senior Research Investigator at Penn Vet’s Comparative Orthopaedic Research Laboratory, and Dr. Dipti Pitta, Assistant Professor in Ruminant Nutrition, will study how healthy microbial communities are influenced by the introduction of bacteria in order to shed new light on the cause of periodontal disease.
 Periodontal disease is a chronic, progressive disease of the gums and periodontal structures that affects one out of every two American adults ages 30 and over. The disease has been associated with other chronic diseases such as cardiovascular disease and arthritis.
Periodontal disease is a chronic, progressive disease of the gums and periodontal structures that affects one out of every two American adults ages 30 and over. The disease has been associated with other chronic diseases such as cardiovascular disease and arthritis.
There is increasing evidence that periodontal disease is associated with oral dysbiosis, a change in the normal microbial population in the mouth. However, it has not been established whether changes in this complex microbial community are a driving cause, or simply a consequence, of the disease.
Schaer and Pitta will profile the bacteria that colonize the mouths of healthy miniature pigs and will compare this to the bacteria found in the mouths of miniature pigs with severe periodontal disease—a process that occurs naturally in these animals at seven to nine years of age. This spontaneous development of disease—and the fact that miniature pigs eat a diet much like humans, chew their food in a way that is similar to humans, and have a dental structure like humans—means that these animals serve as an ideal model for this research.
Using a transplantation model, Schaer and Pitta will discover how healthy microbial communities are influenced by the introduction of oral bacteria from pigs with periodontal disease. Their work would be the first to address a causeeffect relationship between oral bacteria and periodontal disease. Results from this study would lay the groundwork for a translational research platform to explore causal relationships between periodontal disease and degenerative processes in distant organ systems, such as the musculoskeletal and cardiovascular systems.