 María Betina Pampena, a postdoctoral researcher in Michael Betts’s lab at the Perelman School of Medicine, works through the pandemic to identify patterns in patients’ immune responses to COVID-19. The work could lead to more tailored approaches to treatment.
María Betina Pampena, a postdoctoral researcher in Michael Betts’s lab at the Perelman School of Medicine, works through the pandemic to identify patterns in patients’ immune responses to COVID-19. The work could lead to more tailored approaches to treatment.
(Image: Leticia Kuri-Cervantes)
As the rumblings of a pandemic began to be felt at the beginning of the year, scientists at Penn started work to develop a vaccine and assess possible treatments. But the scope of COVID-19 studies at the University goes much broader. Scientists whose typical work finds them investigating autoimmune disease, influenza, HIV/AIDS, Ebola, cancer, hemophilia, and more, are now applying their deep understanding of biology to confront a novel threat.
The more scientists and clinicians observe about the virus, the more avenues of investigation emerge, aiming to shed light on questions such as what happens once the virus enters the body, what treatments might be of benefit, and how society should take action to keep transmission low.
To dig into what scientists around campus are asking and learning, Penn Today spoke with several who have pivoted their research to focus on COVID-19. Their work, while in its early days, is in many cases already finding applications in the fight against this ferocious virus, and may well shape the next steps to defeat it.
What does SARS-CoV-2 do to our lungs?
Another respiratory infection, influenza, has been a focus of research led by Andrew Vaughan of the School of Veterinary Medicine. But Vaughan didn’t hesitate to begin studies of the novel coronavirus once its eventual impact became apparent.
 In the past Andrew Vaughan has studied lung injury and regeneration through the lens of influenza. Now he’s focusing his efforts on understanding how to avoid the damage inflicted by SARS-CoV-2.
In the past Andrew Vaughan has studied lung injury and regeneration through the lens of influenza. Now he’s focusing his efforts on understanding how to avoid the damage inflicted by SARS-CoV-2.
“It’s not a stretch for our lab,” he says. “All the projects in our lab focus on repair and regeneration of the lungs after injury. The majority of my studies are to some degree agnostic about what is causing the injury.”
Earlier work by his group, for example, showed that a lung cell transplant could boost healing in mice affected by a severe bout with flu. Now, graduate students and research specialists in his lab—working no more than two together at a time to maximize social distancing—are conducting new experiments focused more specifically on the biology of SARS-CoV-2, alongside parallel efforts by Edward Morrissey from the Perelman School of Medicine (PSOM). Knowing that the Ace2 receptor on lung cells is the gateway for the virus into the human body, they’re genetically manipulating alveolar type-two lung cells, those that are particularly essential for continuing oxygen exchange deep in the lungs, to alter or block ACE2 gene expression to try to prevent viral entry.
“These alveolar type-two cells seem to be particularly susceptible to injury in both influenza and perhaps even more so in COVID-19,” says Vaughan. “In a perfect world, you might be able to take these genetically edited type-two cells and use them as a cellular therapy. I don’t know that this is going to happen in time to impact this pandemic, but even if the pathogen the next time around is slightly different, we may still be able to employ these types of regenerative responses to help the lung recover better from injury.”
Why are men worse off than women?
In a separate project, Vaughan is partnering with Penn Vet’s Montserrat Anguera to explore a curious feature of COVID-19 disease: the fact that more men than women become severely ill and die. A number of hypotheses have been put forward to explain the disparity, but the two labs are investigating one particular possibility.
“Dr. Anguera had posted something on Twitter saying that the ACE2 gene happens to be on the X chromosome, meaning that women have two copies of it,” says Vaughan. “I immediately texted her and said, ‘I think there’s something to that.’”
Normally women inactivate one of their X chromosomes, but some genes can “escape” this inactivation. This means it’s possible women may have higher ACE2 expression than men. Somewhat counterintuitively, scientists have actually found that higher ACE2 levels actually reduce lung injury, even though ACE2 is also what the virus depends on to enter cells.
Hormone expression levels are, of course, another factor that may influence sex differences in disease. Together, Anguera and Vaughan’s groups are both studying ACE2 expression and exposing alveolar type-two cells to various hormones to see how expression of viral receptors, Ace2 and others, changes. “Ultimately we’d like to see if this changes susceptibility to infection, working with Susan Weiss and others,” says Vaughan.
Do genetics influence susceptibility?
Individual differences in how people respond to infection may be influenced by their unique genomic sequences. Penn Integrates Knowledge Professor Sarah Tishkoff of PSOM and the School of Arts & Sciences, is probing the rich sources of genomic data her group already had in hand to look for patterns that could explain differences in disease susceptibility. As in Vaughan and Anguera’s work, ACE2 is a focus.
 Using genomic samples collected from Africans and those of African ancestry, Sarah Tishkoff is looking for patterns that may
Using genomic samples collected from Africans and those of African ancestry, Sarah Tishkoff is looking for patterns that may
influence disease risk and severity.
“This gene is very important for general health,” Tishkoff says. “Women have two copies, men have one; it plays a role in regulating blood pressure; it’s in the kidneys; it’s in the gut. We want to understand the role that variation at this gene may play in risk for COVID-19, severity of disease in people with underlying health conditions, and differences in the prevalence of disease in men and women.”
Using genomic data from 2,500 Africans collected for another project, Tishkoff’s team is looking for patterns of genetic diversity. Early findings suggest that natural selection may have acted upon on version of the ACE2 gene, making it more common in some African populations with with high exposure to animal viruses.
She’s also collaborating with Anurag Verma and Giorgio Sirugo of Penn Medicine to analyze genetic variation in samples from the Penn Medicine Biobank, looking in particular at people of African descent. “We’re seeing disturbing health disparities with COVID, with African Americans at higher risk for serious illness,” says Tishkoff. “This disparity most likely has to do with inequities in access to health care and socioeconomic factors, but we’re also looking to see if genomic variation may be playing a role.”
Looking ahead, Tishkoff hopes to partner with Daniel Rader and others through the Center for Global Genomics and Health Equity to work with the West Philadelphia community. “We’d like to do testing to understand the prevalence of infection and identify environmental and genetic risk factors for disease,” she says.
How is the immune system reacting?
The immune reaction to SARS-CoV-2 is a double-edged sword. “The immune system is what eliminates the virus,” says E. John Wherry of PSOM. “The immune system is what we need to activate with a good vaccine. But also, especially in many respiratory infections, the immune system is what also causes damage. A healthy outcome means your immune system is striking a balance between killing off the virus and not doing so much damage that it kills you.”
Wherry and PSOM’s Michael Betts have embarked on a study to discern both the magnitude of patients’ immune responses as well as their “flavor,” that is, what components in the immune system are being activated by the coronavirus. They’re doing so by working with clinicians at the Hospital of the University of Pennsylvania (HUP) and, soon, at Penn Presbyterian Medical Center, to collect blood samples from patients with severe and more mild infections, as well as patients who have recovered from illness, to profile their immune reactions.
“We are observing a huge amount of heterogeneity across these patient samples,” says Betts. “But we’re also identifying some relatively unifying characteristics, indicating there are mechanisms that everyone uniformly uses to fight off this infection.”
This variety across patients strongly suggests that the treatments that work for one patient may not for another, Wherry and Betts note. For that reason, they are speaking daily with their colleagues on the front lines of COVID-19 care, relaying what they’re finding out in the lab to adjust and personalize care in the clinic.
“It’s one of the beautiful things about Penn,” says Wherry. “Everyone is working as a team, being selfless, being present, and bringing all their expertise to bear on this crisis.”
What’s the microbiome got to do with it?
Plenty of recent scientific attention has been paid to the role of the gut microbiome in health. But the medical school’s Ronald Collman and Frederic Bushman have been devoting attention to how the community of bacteria, viruses, fungi, and parasites that dwell in the respiratory tract affect health and disease risk. They are now addressing that question in the context of COVID-19.
“There are two reasons we’re interested in studying this,” Collman says. “First is that the microbiome can help set the tone for the immune response to infections, influencing whether a patient ends up with mild or severe disease. And second, the microbiome is where infectious agents that can cause infection can arise from. So if a patient dies of an eventual pneumonia, the pathogen that caused that pneumonia may have been part of that individual’s respiratory tract microbiome.”
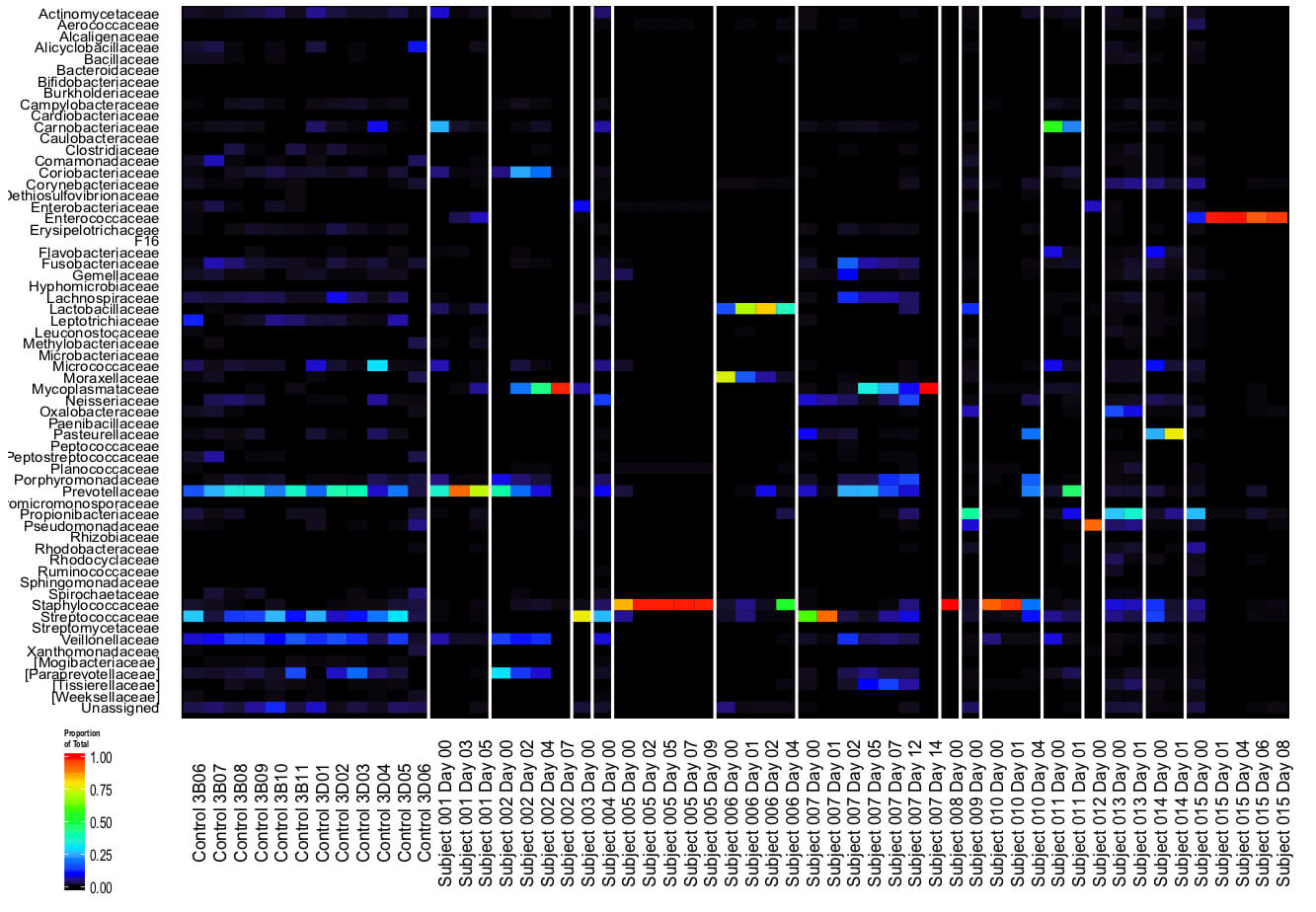 The microbes that live in the respiratory tract may influence disease risk, a focus of attention of earlier studies by Ronald Collman and Frederic Bushman (above). Using samples collected by nurses at HUP, their labs are now looking for patterns in the microbiomes of COVID-19 patients. (Image: Courtesy of the Collman laboratory)
The microbes that live in the respiratory tract may influence disease risk, a focus of attention of earlier studies by Ronald Collman and Frederic Bushman (above). Using samples collected by nurses at HUP, their labs are now looking for patterns in the microbiomes of COVID-19 patients. (Image: Courtesy of the Collman laboratory)
Working with nurses at HUP to collect samples, Collman and Bushman are analyzing the microbiome of both the upper (nose and throat) and lower (lung) portions of the respiratory tract of COVID-19 patients. These samples are being used by other groups, such as those developing diagnostic tests, while Collman and Bushman’s labs work to identify the types and quantities of organisms that compose the microbiome to find patterns in how they correlate with disease.
“We’re hoping that if we can find that the response to the virus is different in people with different upper respiratory tract microbiomes, then we could manipulate the microbiome, using particular antibiotics, for example, to make it more likely that patients would have a mild form of the disease.”
What drugs might make an impact?
Absent a vaccine, researchers are looking to existing drugs—some already approved by the U.S. Food and Drug Administration for other maladies—to help patients recover once infected. Throughout his career, Ronald Harty of Penn Vet has worked to develop antivirals for other infections, such as Ebola, Marburg, and Lassa Fever.
“Our antivirals are sometime referred to as ‘host-oriented’ inhibitors because they’re designed to target the interaction between host and viral proteins,” says Harty. Though many of the biological details of how SARS-CoV-2 interacts with the human body are distinct from the other diseases Harty has studied, his group noticed a similarity: A sequence he’s targeted in other viruses—a motif called PPxY—is also present in the spike protein of SARS-CoV-2, which the coronavirus uses to enter cells.
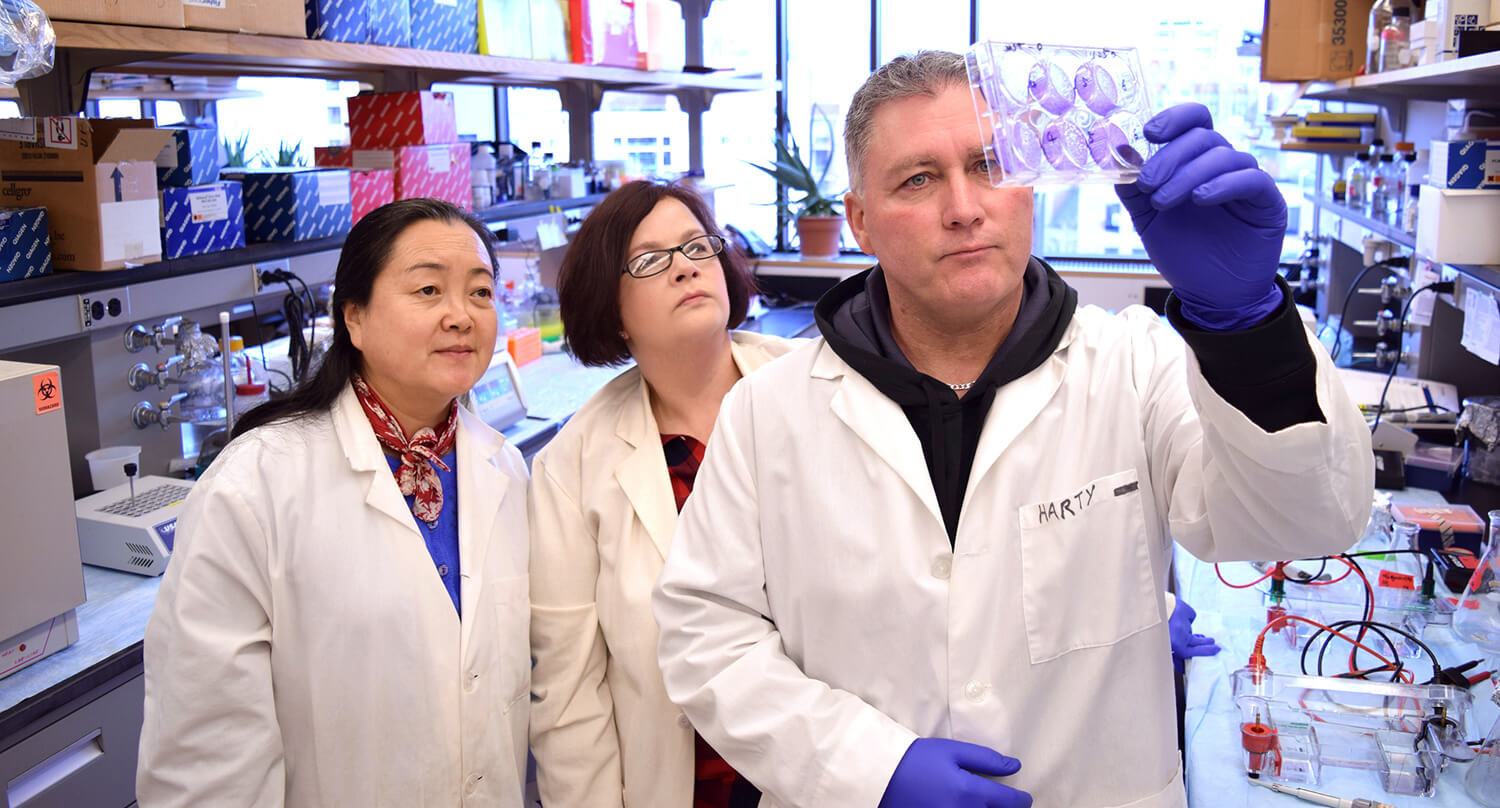 With expertise in analyzing antivirals for diseases like Ebola and Marburg virus, Ronald Harty’s lab is collaborating with other scientists at Penn and elsewhere to determine whether compounds developed to target other diseases may lessen the severity of COVID-19 infections.
With expertise in analyzing antivirals for diseases like Ebola and Marburg virus, Ronald Harty’s lab is collaborating with other scientists at Penn and elsewhere to determine whether compounds developed to target other diseases may lessen the severity of COVID-19 infections.
“This caught our eye,” says Harty, “and piqued our interest in the very intriguing possibility that this PPxY motif could play a role in the severity of this particular virus.”
Harty is testing antivirals he has helped identify that block the replication of Ebola, Marburg, and other viruses to see if they make a dent on the activity of SARS-CoV-2. Those experiments will be done in collaboration with colleagues whose labs can work in BSL-III or -IV laboratories, such as Penn’s Weiss.
Also of interest is the speculation that the coronavirus might disrupt cell-cell junctions in the human body, making them more permeable for virus spread. Harty’s lab will be examining the potential interactions between the viral structural proteins and human proteins responsible for maintaining these cellular barriers.
Another faculty member is assessing whether a drug developed for a very different condition—in this case, pulmonary arterial hypertension (PAH)—could serve coronavirus patients. Henry Daniell of the School of Dental Medicine recently shared news that a drug grown in a plant-based platform to boost levels of ACE2 and its protein product, angiotensin (1-7), was progressing to the clinic to treat PAH. Daniell is now working with Kenneth Margulies from Penn Medicine to explore whether this novel oral therapy can improve the clinical course of patients with symptomatic COVID-19 infection.
 Using his unique plant-based drug development platform, Penn Dental Medicine’s Henry Daniell is working with Kenneth Margulies of Penn Medicine and others to assess whether a novel therapy may help those with serious
Using his unique plant-based drug development platform, Penn Dental Medicine’s Henry Daniell is working with Kenneth Margulies of Penn Medicine and others to assess whether a novel therapy may help those with serious
forms of COVID-19 disease. (Image: Rebecca Elias Abboud)
Reduced ACE2 expression has been linked to acute respiratory distress, severe lung injury, multi-organ failure and death, especially in older patients. The earlier preclinical studies in PAH animal models showed that orally delivered ACE2 made in plant cells accumulated ten times higher in the lungs than in the blood and safely treated PAH. Now, new clinical studies have been developed to explore whether oral supplementation of ACE2 and angiotensin-1-7 can help mitigate complications of COVID-19 disease. The fact that freeze-dried plant cells can be stored at room temperature for as long as a year and can be taken at home by COVID-19 patients make this novel approach an attractive potential option.
“This trial has been given a high priority by the Penn Clinical Trial Working Group,” says Daniell. “I’m pleased that this looks to be on the cusp of moving forward to help the growing number of COVID-19 patients.”
What public health interventions should be taken, and when?
As the coronavirus began to spread in the United States, biologist Joshua Plotkin of the School of Arts & Sciences began to raise alarms about Philadelphia’s St. Patrick’s Day parade, which had been scheduled to be held March 15, potentially drawing thousands to downtown streets. He had good reason to be concerned: His studies of the 1918 flu pandemic had explored disease incidence and spread, and it was hard to avoid noticing the role of the Liberty Loan parade down Broad Street in triggering a rampant spread of flu a century ago.
 Hundreds of thousands of people attended the Liberty Loan parade in Philadelphia in September 1918, an event that contributed to the spread of the Spanish flu. Joshua Plotkin and colleagues at Princeton have shared an analysis of the optimal ways to intervene with public health measures to avoid a serious second peak of cases in the coronavirus pandemic.
Hundreds of thousands of people attended the Liberty Loan parade in Philadelphia in September 1918, an event that contributed to the spread of the Spanish flu. Joshua Plotkin and colleagues at Princeton have shared an analysis of the optimal ways to intervene with public health measures to avoid a serious second peak of cases in the coronavirus pandemic.
Now, with work conducted with two graduate students from Princeton University, Dylan Morris and Fernando Rossine, along with Princeton faculty member Simon Levin, Plotkin has mathematically sound advice for policymakers hoping to effectively stem the spread of a pandemic. In a preprint on arXiv.org, they share “optimal, near-optimal, and robust” strategies for how to time interventions such as social distancing.
“This boils down to knowing what is the best way, of all the infinite possibilities, to intervene using public health measure,” says Plotkin. “That’s a problem we can solve with math, my colleagues Dylan and Fernando realized.”
Their analysis makes the realistic assumption that policymakers can only enforce social distancing for a limited amount of time, and aims to minimize the peak incidence of disease. The optimal strategy, they found, is to start by introducing moderate social distancing measures to keep the incidence rate the same for a period of time. This would mean that every person with COVID-19 would infect one additional person. Then the intervention should switch over to a full suppression—the strongest possible quarantine—for the rest of the period. At the end of that period, all restrictions would be lifted.
“This works because you don’t want to fully suppress disease spread right off the bat,” says Plotkin, “because then at the end, after you remove restrictions, there will be a second peak that is just as large as the first. By employing a moderate suppression at the beginning, you’re building up a population of people who are going to recover and become immune, without letting the epidemic get out of control.”
Unsurprisingly, timing is key. “Attempting the optimal intervention would be disastrous, in practice, because of inevitable errors in timing. Intervening too early is pretty bad, because you get a bigger second peak,” he says. “But intervening too late is even worse. The key lesson is that a robust intervention is more important than an optimal one.”
Plotkin and his colleagues are hoping to share the findings widely, including with local decision makers, to help them navigate a likely second wave of COVID-19.
Montserrat Anguera is an associate professor of biomedical sciences at the University of Pennsylvania School of Veterinary Medicine.
Michael Betts is a professor of microbiology at the University of Pennsylvania Perelman School of Medicine.
Frederic Bushman is the William Maul Measey Professor in Microbiology at the University of Pennsylvania Perelman School of Medicine.
Ronald Collman is a professor of Medicine at the University of Pennsylvania Perelman School of Medicine.
Henry Daniell is vice-chair and W.D. Miller Professor in the Department of Basic and Translational Sciences in the University of Pennsylvania School of Dental Medicine.
Ronald Harty is a professor of pathobiology and microbiology at the University of Pennsylvania School of Veterinary Medicine.
Kenneth Margulies is a professor of medicine and physiology and research and fellowship director of the Heart Failure and Transplant Program at the University of Pennsylvania Perelman School of Medicine.
Joshua Plotkin is the Walter H. and Leonore C. Annenberg Professor of the Natural Sciences in the Department of Biology at the University of Pennsylvania School of Arts & Sciences. He has secondary appointments in the Department of Mathematics and in the School of Engineering and Applied Science’s Department of Computer and Information Science.
Sarah Tishkoff is the David and Lyn Silfen University Professor with appointments in the Perelman School of Medicine’s Department of Genetics and the School of Arts and Sciences’ Department of Biology. A Penn Integrates Knowledge Professor, she is also director of the Penn Center for Global Genomics and Health Equity.
Andrew Vaughan is an assistant professor of biomedical sciences at the University of Pennsylvania School of Veterinary Medicine.
E. John Wherry is chair of the Department of Systems Pharmacology and Translational Therapeutics, director of the Institute for Immunology, and the Richard and Barbara Schiffrin President’s Distinguished Professor at the University of Pennsylvania Perelman School of Medicine.