Research Newsletter
Explore the significant research advancements presented in our Research Newsletter. This collection or research highlights provides valuable insights into our ongoing research efforts and their implications for the field of veterinary science.
Stay connected with all the latest from Penn Vet’s research by signing up for our newsletter.
Previous Issues:
Summer 2024
Discover the research that is making the biggest impact in the latest issue!
Announcements
The new Penn Vet intranet, Inside.Vet, launched on July 25, 2024. Penn Vet community members can access the site via PennKey login.
Please visit the Research section and have a look around. If there are items you would like to see posted there, contact Kathy Kruger, Office of Research and Academic Resources.
New Penn Research Portal
The U@Penn Research Portal has been retired, and the Penn Research Portal has taken its place. The Penn Research Portal is a one-stop shop for information on Animal Operations, Commercialization, Compliance, Finance & Reporting, Pre & Post-Award, Protocol Management, Research Data Management, and Training.
Nature Masterclasses
Nature Masterclasses are professional development training classes for researchers. More than twenty on-demand courses are offered that can be completed on your schedule. Access to the courses is available to anyone with a PennKey.
Looking for Undergraduate Research Assistants?
If you are looking for an undergraduate research assistant or are open to involving an undergraduate in your work, you can post an entry in the Center for Undergraduate Research and Fellowships (CURF) Research Directory. The directory allows interested students to identify and connect with Penn faculty mentors.
Join Penn Vet’s Artificial Intelligence Interest Group
Drs. Fuyu Guan and Darko Stefanovski have started a schoolwide Artificial Intelligence Interest Group. If you are interested in joining the group, please contact Dr. Guan.
FEATURED ARTICLE
Dr. Kotaro Sasaki: Creating the Future of Reproductive Medicine
Kotaro Sasaki, MD, PhD, joined Penn Vet in 2018 and is currently the Richard King Mellon Associate Professor of Biomedical Sciences. His lab is primarily interested in understanding how human urogenital and reproductive organs develop and using this knowledge to develop stepwise and directional induction schemes to generate such organs in vitro from human induced pluripotent stem cells.
Dr. Sasaki became interested in biomedical research in 2001 as a second-year medical student at Hokkaido University when he attended an immunology lecture given by Professor Takashi Nishimura, who fervently explained that through an in-depth understanding of how immune cells work, scientists are now on the cusp of curing cancers through the use of engineered immune cells. Struck by such a transformative idea, Dr. Sasaki joined Dr. Nishimura’s lab as a visiting scholar.
During this period, he discovered that the T cell adhesion molecule very late antigen-2 (VLA-2) is differentially expressed between polarized type-1 and type-2 T cells. After briefly working as an intern in the emergency department of the International Medical Center of Japan, Dr. Sasaki moved to the US and joined the immunology department at the University of Pittsburgh under the supervision of Drs. Walter Storkus and Hideho Okada. As a postdoctoral fellow, he discovered that VLA-4, another T-cell adhesion molecule, and miR-17-92, a micro-RNA cluster, are essential for the homing and activation of therapeutic anti-tumor Type-1 T cells, respectively. This yielded six first-author publications, including publications in Cancer Research and The Journal of Immunology. However, Dr. Sasaki’s interest in clinical research then led him to pursue postgraduate medical training in diagnostic/anatomic pathology at the University of Pittsburgh Medical Center (residency) and the University of Washington Medical Center (renal pathology fellowship). During this time, his detailed histological and immunohistochemical descriptions of various neoplastic and non-neoplastic conditions of urogenital organs, such as adrenal glands and kidneys, nurtured his broader interest in the pathophysiology of the human urogenital organ system.
After completing his clinical training, Dr. Sasaki went back to Japan to pursue his second postdoctoral fellowship in the laboratory of Dr. Mitinori Saitou. There, he studied the development of germ cells in humans and non-human primates using single-cell genomics and stem cell-based approaches. He established robust culture methods to induce human pluripotent stem cells into primordial germ cell (PGC)-like cells, the earliest precursor of both oocytes and sperm, thereby creating a foundation for mechanistic assessment of human germ cells. In parallel, Dr. Sasaki determined that the primate germline originates in the nascent amnion and revealed that germ cell specification pathways of primates and mice have diverged. This discovery finally resolved the mysterious origin of the primate germline and established a framework for the induction of primate germ cells from pluripotent stem cells in vitro.
Human male in vitro gametogenesis
The germ cell lineage ensures transmission of genetic materials to the next generation and, therefore, plays an essential role in the perpetuation of species. Failures in germ cell development result in infertility affecting one in six couples. However, the genetic mechanisms underlying a large fraction of these cases remain elusive due to the lack of tractable models of human germ cell development. Since 2018, the Sasaki lab has focused on establishing new methods to recapitulate human male germ cell development from pluripotent stem cells. First, extending his earlier studies on pluripotent stem cell-derived human PGC-like cells, Dr. Sasaki’s group further induced them to form fetal and adult-stage sperm precursor cells. This platform now allows for mechanistic assessment of human sperm cell development. However, the current protocol only allows differentiation to the spermatocyte stage, and cells do not undergo meiosis to become haploid sperm. The Sasaki lab is currently optimizing this method to allow the successful completion of meiosis, thereby enabling the generation of stem-cell-derived “artificial” sperm cells. This technology will open up a new chapter of reproductive medicine.
Generation of human adrenal gland in dish from pluripotent stem cells
The adrenal cortex is the major endocrine hub for steroid hormone production, and thus regulates a wide array of critical physiologic functions, including immune and stress responses and glucose metabolism. Loss of adrenal function, as occurs in primary adrenal insufficiency, results in a life-threatening disease driven by decreases in cortisol and other critical steroid hormones. To overcome these drawbacks, cell replacement therapy using stem cell-derived adrenal gland has been anticipated. However, because the tissue homeostasis of the adrenal gland is mediated by self-renewal and differentiation of subcortical stem cells, long-term therapeutic effects will require transplantation of both stem cells and surrounding niche cells that support stem cell self-renewal.
Recently, Dr. Sasaki’s team has successfully derived various adrenocortical cell types from human induced pluripotent stem cells using the stepwise and directional induction scheme they invented. Resultant cell types highly resemble normal developmental stages of the human adrenal cortex. Currently, a major goal of the Sasaki lab is to establish self-sustaining and transplantable mature human adrenal cortex organoids that can provide physiologically regulated hormone production for the treatment of primary adrenal insufficiency. As such, their study will not only offer a permanent cure for primary adrenal insufficiency and other adrenal disorders but will also allow for both mechanistic analyses of additional adrenal disorders and for screening therapeutic drugs that regulate adrenal steroidogenesis.
Latest Research
Highlighted News

A hopeful time for Cryptosporidium research (link is external)
Boris Striepen of Penn Vet organized the First Biennial Cryptosporidium Meeting, bringing together researchers and clinicians from around the world to discuss the problems and progress around the parasite and…

Understanding disease prevalence in Pennsylvania wild turkeys (link is external)
Researchers from Penn Vet’s Wildlife Futures Program are collaborating with the Pennsylvania Game Commission and Penn State on a multi-year turkey study.

Frontier Explorers: Penn Vet Scientists on the Cutting Edge
Five of Penn Vet’s newest faculty share their personal passions and what ignites their pursuit of “what’s next” in the profession.
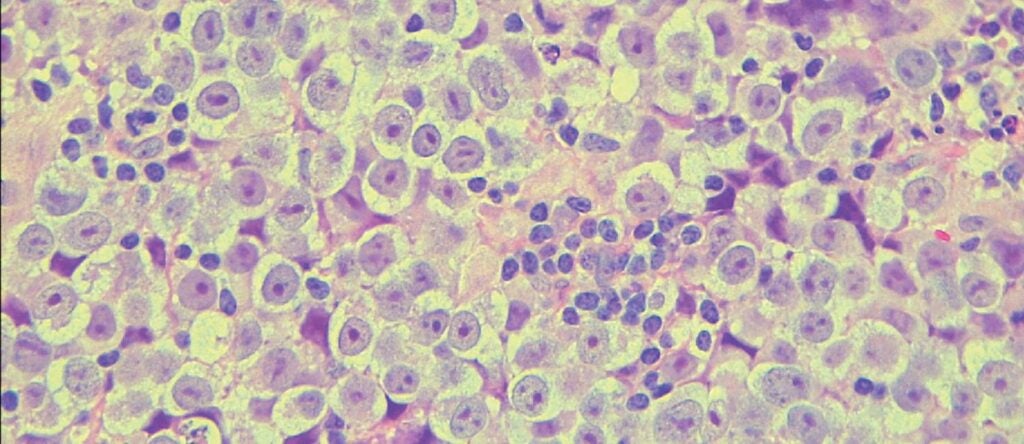
Kotaro Sasaki and his team unveil the genetics of testicular cancer (link is external)
Researchers develop the first in vitro seminoma model, shedding light on chromosomal anomalies and signaling pathways.
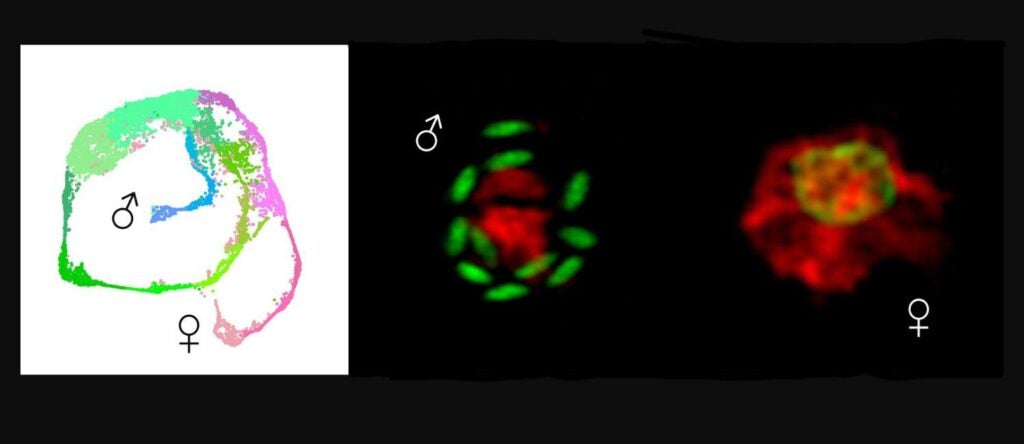
How deadly parasites choose to be male (link is external)
Penn Vet researchers reveal the gene expression across the life cycle of Cryptosporidium and identify the determinant of maleness.

Could we breed cows that emit less methane? (link is external)
In a new study, researchers from the School of Veterinary Medicine identified attributes of low-methane-emitting dairy cows that could be used as targets for selective breeding.
Social interaction is key to survival and reproductive success in primates, including humans. Optimizing outcomes from these encounters requires a calculated approach to cooperation and competition—knowing whom to trust, whom to avoid, or whom to confront confers an evolutionary advantage.
Penn Vet’s Division of Experimental Retinal Therapies (ExpeRTs), led by Gustavo D. Aguirre, VMD, PhD, and William A. Beltran, DVM, PhD, was well represented at this year’s Annual Meeting of the Association for Research in Vision and Ophthalmology (ARVO), held in Seattle, Washington, May 5—9, 2024. Nine abstracts were presented by the ExpeRTs.
Clinical characterization of canine cone-rod dystrophy with early foveo-macular degeneration.
G.D. Aguirre, V.L. Dufour, L. Murgiano, J.C. Kwok, W.A. Beltran.
Adaptive glial cell and gene expression alterations in retinas of treated and untreated RPE65 mutant dogs.
T. Appelbaum, E. Santana, D. Smith, W.A. Beltran, G.D. Aguirre.
Development of OPGx-BEST1 AAV gene therapy for the treatment of bestrophinopathies.
M. Choudhary, A. Gray, Y. Sato, J.C. Kwok, V. Arora, B.R. Yerxa, A.V. Cideciyan, G.D. Aguirre, W.A. Beltran, A. Jayagopal.
A canine model for ABCA4 Stargardt disease: in-vivo phenotypic characterization and natural disease history.
V.L. Dufour, A.V. Cideciyan, J. Kwok, A. Gray, Y. Sato, K. Quigley, W.A. Beltran, G.D. Aguirre.
Assessment of the Orbit™ Subretinal Delivery System (OSDS™) and prototypes in adult and juvenile canine eyes.
J.C. Kwok, A. Gray, Y. Sato, S. Reichenbacker, T. Meyer, K. Stoner, W.A. Beltran.
Frameshift variant in the Cirneco dell’Etna dog with syndromic retinopathy and tremors.
L. Murgiano, J.K. Niggel, L. Benedicenti, A. Bionda, M. Cortellari, P. Crepaldi, G.K. Aguirre, W.A. Beltran, G.D. Aguirre.
DNA extraction and amplification from osmicated EPONbedded retinal and corneal tissues.
J. Niggel, G. Aguirre, L. Murgiano.
Identification of photoreceptor-specific promoters for gene therapy at advanced stages of retinal degeneration.
R. Sudharsan, L. Murgiano, A. Ahuja, Y. Sato, J.C. Kwok, M. Sedorovitz, G.D. Aguirre, L. Byrne, W.A. Beltran.
Ultrastructure expansion microscopy for characterization of the canine photoreceptor sensory cilium.
K. Takahashi, R. Sudharsan, W.A. Beltran.
Honors & Achievements
Dr. Kotaro Sasaki Named Richard King Mellon Associate Professor
Kotaro Sasaki, MD, PhD, has been named the Richard King Mellon Associate Professor of Biomedical Sciences. Dr. Sasaki’s research is focused on the development and pathophysiology of urogenital and reproductive organ systems; his laboratory is advancing the understanding of human infertility, reproduction, and endocrinology. He possesses an exceptional record of academic accomplishments.
Dr. Louise H. Moncla Named Penn Vet’s First Pew Biomedical Scholar
The Pew Charitable Trusts has named Louise H. Moncla, PhD, a 2024 Pew Scholar in the Biomedical Sciences. Dr. Moncla is Penn Vet’s first Pew Scholar. Her research will reconstruct how avian influenza viruses accumulate mutations that allow them to jump from birds to humans.
Dr. Boris Striepen Receives MERIT Award from the NIAID
Boris Striepen, PhD, achieved the exceptional honor of receiving a Method to Extend Research in Time (MERIT) Award from the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH). A MERIT Award is provided to support the principal scientific endeavor of an investigator who has demonstrated superior competence and outstanding productivity. These awards enable NIH institutes to give investigators with stellar records of research accomplishment a five-year award with the possibility of extending the initial award for up to five additional years without the usual competitive review. This award will support Dr. Striepen’s Genetic Analysis of Cryptosporidium project, which was first supported in 2014, and is now in its third round of funding. This project produced a breakthrough in genetic engineering for the Cryptosporidium parasite and several other key advances.
Dr. Dipti Pitta Receives More than Half a Million Dollars to Study Reducing Methane Emissions in Dairy Cattle
Dipti Pitta, MVSc, PhD, has received a $508,884 grant from Gerstner Philanthropies to support her continuing work to imprint the rumen function and prevent methanogen colonization in dairy calves, with the potential of permanently curbing methane emissions from dairy herds in the United States. This study represents a vital step towards developing novel microbial interventions to not only curb methane emissions but also to improve the performance of the American dairy system and the sustainability of animal agriculture.
Dr. Stephen Cole Presents to Presidential Advisory Council on Antimicrobial Resistance
Stephen D. Cole, VMD, presented his work on carbapenem-resistant organisms in companion animals at a meeting of the Presidential Advisory Council on Combating Antimicrobial-Resistant Bacteria (PACCARB) on May 21, 2024. The PACCARB is a federal advisory council that provides advice, information, and recommendations on all things related to antimicrobial resistance to the Secretary of Health and Human Services (HHS) in close consultation with the Departments of Agriculture (USDA) and Defense (DoD). Federal advisory councils are a way for the federal government to use the knowledge of non-federal subject matter experts to help support and inform federal policies and actions in an open and public forum.
Dr. Sue McDonnell Receives 2024 NAEAA Don Henneke Educational Impact Award
The National Association of Equine Affiliated Academics (NAEAA) has named Sue McDonnell, PhD,adjunct professor of reproductive behavior and founding head of Penn Vet’s equine behavior program, the 2024 recipient of the Don Henneke Educational Impact Award.
Dr. Nicola Mason Receives Alan Kelly International Achievement Award
Nicola J. Mason, BVetMed, PhD, is this year’s recipient of the prestigious Alan Kelly International Achievement Award, which is presented to an individual who is currently involved in world-class innovation in canine health and welfare research. The $100K prize is intended to finance future projects and to reward the winner’s outstanding contribution to canine health and welfare. The award is part of The International Canine Health Awards, which are granted by The Kennel Club Charitable Trust.
PETRL Receives Full Re-Accreditation by A2LA
The Pennsylvania Equine Toxicology and Research Laboratory (PETRL), directed by Mary A. Robinson, VMD, PhD, has been fully re-accredited by the American Association for Laboratory Accreditation (A2LA) to ISO/IEC 17025 to perform chemical testing. In addition, characterization/identification of unknown samples in a variety of matrices has been added to the laboratory’s scope of accreditation. The accreditation is valid June 5, 2024 — May 31, 2026.
Established in 1978, A2LA is among the largest accreditation bodies in the world and the only independent, 501(c)3, non-profit, internationally recognized accreditation body in the United States that offers a full range of comprehensive conformity assessment accreditation services.
Dr. Chris Hunter Delivers Distinguished Lecture at Immunology2024
Christopher A. Hunter, BSc, PhD, delivered a Distinguished Lecture at Immunology2024, the annual meeting of the American Association of Immunologists, on May 4, 2024, in Chicago, Illinois. Dr. Hunter’s presentation was entitled, From parasites to mRNA vaccines: cytokine networks that dictate T cell tempo.
Dr. Lauren Powell Receives the Animals Young Investigator Award
Lauren R. Powell, PhD, is the recipient of the 2023 Animals Young Investigator Award. Animals is a PubMed-indexed, international, peer-reviewed journal affiliated with the World Association of Zoos and Aquariums (WAZA), European College of Animal Welfare and Behavioural Medicine (ECAWBM), and the Federation of European Laboratory Animal Science Associations (FELASA). The Young Investigator Award was established to acknowledge the achievements of young investigators in different fields of study involving animals, including zoology, ethnozoology, animal science, animal ethics, and animal welfare.
Dr. Katie Bardales Receives ACVIM Resident Research Abstract Award
Kathleen Bardales, DVM, is the recipient of an ACVIM Resident Research Abstract Award for her abstract, Transcriptional profiling of the immune tumor microenvironment in canine cutaneous mast cell tumors. The award was presented at the 2024 ACVIM Forum in Minneapolis. Dr. Bardales is mentored by Drs. Matthew J. Atherton, Jennifer Lenz, and Charles-Antoine Assenmacher.
Dr. Jared Crofts Receives JAVMA Journal Award
Jared L. Crofts, DVM, received a 2024 JAVMA Interns Journal Award for his publication, Increased incidence and shift in the location of gunshot wound injuries in dogs and cats during the COVID-19 pandemic. Co-authors include members of Dr. Crofts’ Emergency and Critical Care team, which was led by Erica L. Reineke, VMD.
Dr. Rebecka Hess Has Joined the Center for Clinical Epidemiology and Biostatistics
Rebecka S. Hess, MSCE, DVM, Dipl. ACVIM, has joined Penn’s Center for Clinical Epidemiology and Biostatistics (CCEB). The CCEB is an interdisciplinary and interdepartmental program that links clinical epidemiology and biostatistics within the Perelman School of Medicine, the University of Pennsylvania Health System, and the Penn community. The Center exists to improve the health of individuals, communities, and populations through rigorous, innovative research and training.

Steps to Cure Sarcoma 5K Raises More than $250,000 for Research
The 10th annual Steps to Cure Sarcoma fundraiser was held at Wilson Farm Park in Wayne, PA, on Sunday, May 19, 2024. Initiated in 2014, the 5K walk/run has raised more than $1 million for basic and translational sarcoma research being performed at the Schools of Medicine and Veterinary Medicine at the University of Pennsylvania, as well as at the Children’s Hospital of Philadelphia. Runners and walkers from Penn Vet’s Comparative Immunotherapy Program team, the Canine Cancer Crushers, joined more than 50 other teams and collectively raised more than $250,000 for sarcoma research! Two canine cancer immunotherapy projects are being supported by the sarcoma program at Penn, and will hopefully lead to the implementation of two clinical trials in dogs with osteosarcoma that will benefit both canine and human patients.
Select Publications and Grants
Read the latest scholarly publication and grant information from Penn Vet researchers.
Aldridge DL, Moodley D, Park J, Phan AT, Rausch M, White KF, Ren Y, Golin K, Radaelli E, Kedl R, Holland PM, Hill J, and Hunter CA. Endogenous IL-27 during toxoplasmosis limits early monocyte responses and their inflammatory activation by pathological T cells. Mbio 2024;15. https://journals.asm.org/doi/10.1128/mbio.00083-24
Andrews C, Aronson L, Church M, and Piegols H. Dorsal rhinotomy in a dog with a chondro-osseous respiratory epithelial adenomatoid hamartoma: a case report. J Am Vet Med Assoc 2024;262:1-4. https://doi.org/10.2460/javma.23.10.0559
Araos J, Driessen B, Brandly J, Gorenberg E, Heerdt P, Bruhn A, Martin-Flores M, Adler A, and Hopster K. Optimization of lung ventilation and perfusion in anesthetized horses using a ventilation mode with flow-limited expiration. American Journal of Veterinary Research 2024;85. https://doi.org/10.2460/ajvr.23.09.0200
Avison A, Gelzer AR, Reef VB, Wulster Bills KB, de Solis CN, Kraus MS, Slack J, Stefanovski D, Deacon LJ, and Underwood C. Twenty-four hour continuous transvenous temporary right ventricular pacing in healthy horses. J Vet Intern Med 2024;38:1751-1764. https://onlinelibrary.wiley.com/doi/10.1111/jvim.17027
Baker L, Bender J, Ferguson J, Rassler S, Pitta D, Chann S, and Dou ZX. Leveraging dairy cattle to upcycle culled citrus fruit for emission mitigation and resource co-benefits: A case study. Resources Conservation and Recycling 2024;203. https://doi.org/10.1016/j.resconrec.2024.107452
Banse HE, Kedrowicz A, Michel KE, Burton EN, Yvorchuk-St Jean K, Anderson J, Anderson S, Barr MC, Boller E, Chaney K, Dyer Inzana K, Matthew SM, Rollins D, Salisbury SK, Schmidt P, Smith N, and Trace C. Implementing Competency-Based Veterinary Education: A Survey of AAVMC Member Institutions on Opportunities, Challenges, and Strategies for Success. Journal of Veterinary Medical Education 2024;51:155-163. https://utppublishing.com/doi/10.3138/jvme-2023-0012
Benedetti F, Silvestri G, Denaro F, Finesso G, Contreras-Galindo R, Munawwar A, Williams S, Davis H, Bryant J, Wang Y, Radaelli E, Rathinam CV, Gallo RC, and Zella D. Mycoplasma DnaK expression increases cancer development in vivo upon DNA damage. Proceedings of the National Academy of Sciences of the United States of America 2024;121. https://www.pnas.org/doi/10.1073/pnas.2320859121
Box JR, Oyama MA, Mosenco AS, and Hess RS. Effect of sodium-glucose cotransporter 2 inhibitor canagliflozin on interstitial glucose concentration in insulin-treated diabetic dogs. J Vet Intern Med 2024;38:1353-1358. https://onlinelibrary.wiley.com/doi/10.1111/jvim.17053
Brown KA, Gregorio EN, Barot D, Usimaki A, Linardi RL, Missanelli JR, You YW, Robinson MA, and Ortved KF. Single-dose nonsteroidal anti-inflammatory drugs in horses have no impact on concentrations of cytokines or growth factors in autologous protein solution and platelet-rich plasma. American Journal of Veterinary Research 2024;85. https://doi.org/10.2460/ajvr.23.11.0258
Carlson JA, Shetye SS, Sun M, Weiss SN, Birk DE, and Soslowsky LJ. Collagen V haploinsufficiency in female murine patellar tendons results in altered matrix engagement and cellular density, demonstrating decreased healing. J Orthop Res 2024;42:950-960. https://onlinelibrary.wiley.com/doi/10.1002/jor.25740
Carvalho AM, Costa RS, Lago A, Bacellar O, Beiting DP, Scott P, Carvalho LP, and Carvalho EM. In Situ versus Systemic Immune Response in the Pathogenesis of Cutaneous Leishmaniasis. Pathogens 2024;13. https://www.mdpi.com/2076-0817/13/3/199
Chong ACN, Vandana JJ, Jeng G, Li G, Meng Z, Duan X, Zhang T, Qiu Y, Duran-Struuck R, Coker K, Wang W, Li Y, Min Z, Zuo X, de Silva N, Chen Z, Naji A, Hao M, Liu C, and Chen S. Checkpoint kinase 2 controls insulin secretion and glucose homeostasis. Nat Chem Biol 2024;20:566-576. https://doi.org/10.1038/s41589-023-01466-4
Cortellini S, DeClue AE, Giunti M, Goggs R, Hopper K, Menard JM, Rabelo RC, Rozanski EA, Sharp CR, Silverstein DC, Sinnott-Stutzman V, and Stanzani G. Defining sepsis in small animals. J Vet Emerg Crit Care (San Antonio) 2024;34:97-109. https://onlinelibrary.wiley.com/doi/10.1111/vec.13359
Cournoyer A, Amerman H, Assenmacher CA, Durham A, Perry JA, Gedney A, Keuler N, Atherton MJ, and Lenz JA.Quantification of CD3, FoxP3, and granzyme B immunostaining in canine renal cell carcinoma. Vet Immunol Immunopathol 2024;271:110741. https://doi.org/10.1016/j.vetimm.2024.110741
Craven A, Todd-Donato A, Stokol T, Liepman R, Glasberg I, Wilkins P, Luethy D, Wong D, Schoster A, van den Brom-Spierenburg AJ, and Tomlinson JE. Clinical findings and outcome predictors for multinodular pulmonary fibrosis in horses: 46 cases (2009-2019). J Vet Intern Med 2024;38:1842-1857. https://onlinelibrary.wiley.com/doi/10.1111/jvim.17084
Crockett AM, Kebir H, Anderson SA, Jyonouchi S, Romberg N, and Alvarez JI. 22q11.2 Deletion-Associated Blood-Brain Barrier Permeability Potentiates Systemic Capillary Leak Syndrome Neurologic Features. J Clin Immunol 2024;44:87. https://doi.org/10.1007/s10875-024-01686-w
Dey N, Koumenis C, Ruggero D, Fuchs SY, and Diehl JA. miR-217 Regulates Normal and Tumor Cell Fate Following Induction of Endoplasmic Reticulum Stress. Mol Cancer Res 2024;22:360-372. https://aacrjournals.org/mcr/article/22/4/360/741846/miR-217-Regulates-Normal-and-Tumor-Cell-Fate
Didehvar DS, Lanza MR, Atherton MJ, and Lenz JA. Malignant transformation and subsequent leptomeningeal carcinomatosis of a gastric polyp in a dog. J Vet Intern Med 2024;38:1744-1750. https://onlinelibrary.wiley.com/doi/10.1111/jvim.17072
Didier A, Bourner M, Kleks G, Zolty A, Kumar B, Nichols T, Durynski K, Bender S, Gibison M, Murphy L, Ellis JC, Dong DW, and Kashina A. Prospective fecal microbiomic biomarkers for chronic wasting disease. Microbiol Spectr 2024;12:e0375022. https://journals.asm.org/doi/10.1128/spectrum.03750-22
DiGuilio KM, Del Rio EA, Harty RN, and Mullin JM. Micronutrients at Supplemental Levels, Tight Junctions and Epithelial Barrier Function: A Narrative Review. International Journal of Molecular Sciences 2024;25. https://www.mdpi.com/1422-0067/25/6/3452
DiNinni A, and Hess RS. Development of a requirement for exogenous insulin treatment in dogs with hyperglycemia. J Vet Intern Med 2024;38:980-986. https://onlinelibrary.wiley.com/doi/10.1111/jvim.16990
Doshi BS, Samelson-Jones BJ, Nichols TC, Merricks EP, Siner JL, French RA, Lee BJ, Arruda VR, and Callan MB. AAV gene therapy in companion dogs with severe hemophilia: Real-world long-term data on immunogenicity, efficacy, and quality of life. Mol Ther Methods Clin Dev 2024;32:101205. https://doi.org/10.1016/j.omtm.2024.101205
Dou Z, Dierenfeld ES, Wang X, Chen X, and Shurson GC. A critical analysis of challenges and opportunities for upcycling food waste to animal feed to reduce climate and resource burdens. Resources, Conservation and Recycling 2024;203. https://doi.org/10.1016/j.resconrec.2024.107418
Dreese K, Murphy K, Burke J, and Silverstein DC. Seizures in 3 juvenile dogs after intravenous anesthetic drug withdrawal during weaning from mechanical ventilation suspected to be a sign of iatrogenic withdrawal syndrome. Journal of Veterinary Emergency and Critical Care 2024;34:173-178. https://onlinelibrary.wiley.com/doi/10.1111/vec.13366
Duan J, Keeler E, McFarland A, Scott P, Collman RG, and Bushman FD. The virome of the kitome: small circular virus-like genomes in laboratory reagents. Microbiol Resour Announc 2024;13:e0126123. https://journals.asm.org/doi/10.1128/mra.01261-23
Ede T, Ceribelli M, and Parsons TD. Gilts prefer an open pen to a stall. Sci Rep 2024;14:9684. https://doi.org/10.1038/s41598-024-60617-2
Ethgen LM, Pastore C, Lin C, Reed DR, Hung LY, Douglas B, Sinker D, Herbert DR, and Belle NM. A Trefoil factor 3-Lingo2 axis restrains proliferative expansion of type-1 T helper cells during GI nematode infection. Mucosal Immunol 2024;17:238-256. https://doi.org/10.1016/j.mucimm.2024.02.003
Facka A, Frair J, Keller T, Miller E, Murphy L, and Ellis JC. Spatial patterns of anticoagulant rodenticides in three species of medium-sized carnivorans in Pennsylvania. Canadian Journal of Zoology 2024;102:443-454. https://cdnsciencepub.com/doi/10.1139/cjz-2023-0131
Fairfield DK, Singh A, Hawker W, Richardson D, Mayhew P, Balsa I, Culp WTN, Cinti F, Buote NJ, Massari F, Griffin MA, Gibson E, Runge JJ, and Chanoit G. Minimally invasive splenectomy is associated with a low perioperative complication rate and short operative time in cats. Journal of the American Veterinary Medical Association 2024;262:1-7. https://doi.org/10.2460/javma.23.11.0616
Furey C, Scher G, Ye N, Kercher L, DeBeauchamp J, Crumpton JC, Jeevan T, Patton C, Franks J, Rubrum A, Alameh MG, Fan SHY, Phan AT, Hunter CA, Webby RJ, Weissman D, and Hensley SE. Development of a nucleoside-modified mRNA vaccine against clade 2.3.4.4b H5 highly pathogenic avian influenza virus. Nat Commun 2024;15:4350. https://doi.org/10.1038/s41467-024-48555-z
Gibson EA, and Culp WTN. Canine Prostate Cancer: Current Treatments and the Role of Interventional Oncology. Vet Sci2024;11. https://www.mdpi.com/2306-7381/11/4/169
Gorr SC, Leeb C, Zollitsch W, Winckler C, and Parsons TD. Ad libitum feeding systems for lactating sows: effects on productivity and welfare of sows and piglets. Animal 2024;18:101093. https://doi.org/10.1016/j.animal.2024.101093
Gray SM, Goonatilleke E, Emrick MA, Becker JO, Hoofnagle AN, Stefanovski D, He W, Zhang G, Tong J, Campbell J, and D’Alessio DA. High Doses of Exogenous Glucagon Stimulate Insulin Secretion and Reduce Insulin Clearance in Healthy Humans. Diabetes 2024;73:412-425. https://diabetesjournals.org/diabetes/article/73/3/412/153920/High-Doses-of-Exogenous-Glucagon-Stimulate-Insulin
Greening SS, Haman K, Drazenovich T, Chacon-Heszele M, Scafini M, Turner G, Huckabee J, Leonhardt J, vanWestrienen J, Perelman M, Thompson P, and Keel MK. Validation of a Field-Portable, Handheld Real-Time PCR System for Detecting Pseudogymnoascus destructans, the Causative Agent of White-Nose Syndrome in Bats. J Wildl Dis 2024;60:298-305. https://doi.org/10.7589/JWD-D-23-00083
Hickok NJ, Li BY, Oral E, Zaat SAJ, Armbruster DA, Atkins GJ, Chen AF, Coraca-Huber DC, Dai TH, Greenfield EM, Kasinath R, Libera M, Marques CNH, Moriarty TF, Phillips KS, Raghuraman K, Ren DC, Rimondini L, Saeed K, Schaer TP, Schwarz EM, Spiegel C, Stoodley P, Truong VK, Tsang STJ, Wildemann B, Zelmer AR, and Zinkernagel AS. The 2023 Orthopedic Research Society’s international consensus meeting on musculoskeletal infection: Summary from the in vitro section. Journal of Orthopaedic Research 2024;42:512-517. https://onlinelibrary.wiley.com/doi/10.1002/jor.25774
Huh T, Achilles EJ, Massey LK, Loughran KA, Larouche-Lebel E, Convey V, McKaba VF, Crooks A, Kraus MS, Gelzer AR, and Oyama MA. Utility of focused cardiac ultrasonography training in veterinary students to differentiate stages of subclinical myxomatous mitral valve disease in dogs. J Vet Intern Med 2024;38:1325-1333. https://onlinelibrary.wiley.com/doi/10.1111/jvim.17056
Jeong S, Franklin SH, van Eps AW, Lean N, and Ahern BJ. Implementation of a prototype dynamic laryngoplasty system in standing sedated horses provided arytenoid abduction control at seven days postoperatively. American Journal of Veterinary Research 2024;85. https://doi.org/10.2460/ajvr.23.11.0256
Kim U, Debnath R, Maiz JE, Rico J, Sinha S, Blanco MA, and Chakrabarti R. ΔNp63 regulates MDSC survival and metabolism in triple-negative breast cancer. iScience 2024;27. https://doi.org/10.1016/j.isci.2024.109366
Leduc L, Underwood C, Stefanovski D, Hurcombe S, and Navas de Solis C. Evaluation of remote assistance for point-of-care ultrasonography in a large animal hospital: a controlled randomized trial. J Am Vet Med Assoc 2024;262:680-684. https://doi.org/10.2460/javma.23.09.0511
Li C, Powell L, Stamatakis E, McGreevy P, Podberscek A, Bauman A, and Edwards K. Is dog walking suitable for physical activity promotion? Investigating the exercise intensity of on-leash dog walking. Preventive Medicine Reports 2024;41. https://doi.org/10.1016/j.pmedr.2024.102715
Maddock KJ, Burbick CR, Cole SD, Daniels JB, LeCuyer TE, Li XZ, Loy JD, Sanchez S, Stenger BLS, and Diaz-Campos D. A One Health perspective on the use of genotypic methods for antimicrobial resistance prediction. J Am Vet Med Assoc 2024;262:303-312. https://doi.org/10.2460/javma.23.12.0687
Mallikarjun A, Shroads E, and Newman RS. Perception of vocoded speech in domestic dogs. Anim Cogn 2024;27:34. https://doi.org/10.1007/s10071-024-01869-3
Matsuda R, Sorobetea D, Zhang JN, Peterson ST, Grayczyk JP, Yost W, Apenes N, Kovalik ME, Herrmann B, O’Neill RJ, Bohrer AC, Lanza MT, Assenmacher CA, Mayer-Barber KD, Shin S, and Brodsky IE. A TNF-IL-1 circuit controls within intestinal pyogranulomas. Journal of Experimental Medicine 2024;221. https://rupress.org/jem/article/221/3/e20230679/276568/A-TNF-IL-1-circuit-controls-Yersinia-within
McClosky M, Cole S, Seidel EJ, and Hess RS. Clinical Differences in Dogs with Enterococcal Bacteriuria Compared with Other Bacteriuria: A Retrospective, Case-Control Study. J Am Anim Hosp Assoc 2024;60:53-59. https://meridian.allenpress.com/jaaha/article-abstract/60/2/53/499141/Clinical-Differences-in-Dogs-with-Enterococcal?redirectedFrom=fulltext
Mistrick J, Kipp EJ, Weinberg SI, Adams CC, Larsen PA, and Craft ME. Microbiome diversity and zoonotic bacterial pathogen prevalence in Peromyscus mice from agricultural landscapes and synanthropic habitat. Mol Ecol 2024;33:e17309. https://onlinelibrary.wiley.com/doi/10.1111/mec.17309
Mitre E, Kameni M, Musaigwa F, Kamguia LM, Kamdem SD, Mbanya G, Lamberton PHL, and Komguep Nono J. Harnessing Schistosoma-associated metabolite changes in the human host to identify biomarkers of infection and morbidity: Where are we and what should we do next? PLOS Neglected Tropical Diseases 2024;18. https://doi.org/10.1371/journal.pntd.0012009
Mizukami K, Dorsey-Oresto A, Raj K, Eringis A, Furrow E, Martin E, Yamanaka D, Kehl A, Kolicheski A, Jagannathan V, Leeb T, Lionakis MS, and Giger U. Increased susceptibility to Mycobacterium avium complex infection in miniature Schnauzer dogs caused by a codon deletion in CARD9. Sci Rep 2024;14:10346. https://doi.org/10.1038/s41598-024-61054-x
Montero D, Torrecillas S, Serradell A, Nedoluzhko A, Fernández-Montero A, Makol A, Monzón-Atienza L, Valdenegro V, Sanahuja I, Galindo-Villegas J, and Acosta F. Phytogenics enhance welfare and vaccine efficacy against in European seabass juveniles. Aquaculture 2024;585. https://doi.org/10.1016/j.aquaculture.2024.740714
Moriarty TF, Hickok NJ, Saeed K, Schaer TP, Chen AF, and Schwarz EM. The 2023 Orthopaedic Research Society International Consensus Meeting on musculoskeletal infection. J Orthop Res 2024;42:497-499. https://onlinelibrary.wiley.com/doi/10.1002/jor.25714
Mosala P, Mpotje T, Aziz NA, Ndlovu H, Musaigwa F, Nono JK, and Brombacher F. Cysteinyl leukotriene receptor-1 as a potential target for host-directed therapy during chronic schistosomiasis in murine model. Frontiers in Immunology 2024;15. https://doi.org/10.3389/fimmu.2024.1279043
Mwakibete L, Greening SS, Kalantar K, Ahyong V, Anis E, Miller EA, Needle DB, Oglesbee M, Thomas WK, Sevigny JL, Gordon LM, Nemeth NM, Ogbunugafor CB, Ayala AJ, Faith SA, Neff N, Detweiler AM, Baillargeon T, Tanguay S, Simpson SD, Murphy LA, Ellis JC, Tato CM, and Gagne RB. Metagenomics for Pathogen Detection During a Mass Mortality Event in Songbirds. J Wildl Dis 2024;60:362-374. https://doi.org/10.7589/JWD-D-23-00109
Nolen-Walston RD, Stefanoski D, and Conrado FO. Association of leukergy (in vitro leukocyte aggregation) with systemic inflammation in dogs and cats (2017-2022). Veterinary Clinical Pathology 2024;53:40-46. https://onlinelibrary.wiley.com/doi/10.1111/vcp.13325
Olsson IAS, Powell L, Ackerman R, Reinhard CL, Serpell J, and Watson B. A prospective study of mental wellbeing, quality of life, human-animal attachment, and grief among foster caregivers at animal shelters. Plos One 2024;19. https://doi.org/10.1371/journal.pone.0301661
Palmisano M, Kulp J, Bender S, Stefanovski D, Robinson M, and Johnson A. Measurement of 8-hydroxy-2′-deoxyguanosine in serum and cerebrospinal fluid of horses with neuroaxonal degeneration and other causes of proprioceptive ataxia. Journal of Veterinary Internal Medicine 2024;38:1207-1213. https://onlinelibrary.wiley.com/doi/10.1111/jvim.16988
Pan SY, Souza LAC, Worker CJ, Mendez MER, Gayban AJB, Cooper SG, Solano AS, Bergman RN, Stefanovski D, Morton GJ, Schwartz MW, and Earley YF. (Pro)renin receptor signaling in hypothalamic tyrosine hydroxylase neurons is required for obesity-associated glucose metabolic impairment. Jci Insight 2024;9. https://doi.org/10.1172/jci.insight.174294
Pardy RD, Walzer KA, Wallbank BA, Byerly JH, O’Dea KM, Cohn IS, Haskins BE, Roncaioli JL, Smith EJ, Buenconsejo GY, Striepen B, and Hunter CA. Analysis of intestinal epithelial cell responses to highlights the temporal effects of IFN-γ on parasite restriction. Plos Pathogens 2024;20. https://doi.org/10.1371/journal.ppat.1011820
Perrone J, Haroz R, D’Orazio J, Gianotti G, Love J, Salzman M, Lowenstein M, Thakrar A, Klipp S, Rae L, Reed MK, Sisco E, Wightman R, and Nelson LS. National Institute on Drug Abuse Clinical Trials Network Meeting Report: Managing Patients Exposed to Xylazine-Adulterated Opioids in Emergency, Hospital and Addiction Care Settings. Ann Emerg Med 2024;84:20-28. https://doi.org/10.1016/j.annemergmed.2024.01.041
Pletcher JS, Zimmer JL, Liu CC, Beierschmitt A, and Lewin AC. Ocular examination findings and selected ophthalmic diagnostic tests in African green monkeys (Chlorocebus aethiops sabaeus). Vet Ophthalmol 2024;27:158-169. https://onlinelibrary.wiley.com/doi/10.1111/vop.13132
Prasasya RD, Caldwell BA, Liu Z, Wu S, Leu NA, Fowler JM, Cincotta SA, Laird DJ, Kohli RM, and Bartolomei MS. Iterative oxidation by TET1 is required for reprogramming of imprinting control regions and patterning of mouse sperm hypomethylated regions. Developmental Cell 2024;59:1010-1027.e1018. https://doi.org/10.1016/j.devcel.2024.02.012
Raftery K, Rahman T, Smith N, Schaer T, and Newell N. The role of the nucleus pulposus in intervertebral disc recovery: Towards improved specifications for nucleus replacement devices. J Biomech 2024;166:111990. https://doi.org/10.1016/j.jbiomech.2024.111990
Rissi DR, Miller AD, Demeter EA, Church ME, and Koehler JW. Diagnostic immunohistochemistry of primary and secondary central nervous system neoplasms of dogs and cats. J Vet Diagn Invest 2024;36:153-168. https://journals.sagepub.com/doi/10.1177/10406387231221858
Sacramento LA, Amorim CF, Lombana CG, Beiting D, Novais F, Carvalho LP, Carvalho EM, and Scott P. CCR5 promotes the migration of pathological CD8 T cells to the leishmanial lesions. Plos Pathogens 2024;20. https://doi.org/10.1371/journal.ppat.1012211
Sannajust K, Palmisano M, Whisenant K, Slack J, Holt D, Woodrow J, and Manzi TJ. Computed tomographic diagnosis of a common bile duct abscess and secondary extrahepatic biliary duct obstruction in a potbelly pig. Vet Radiol Ultrasound2024;65:303-307. https://onlinelibrary.wiley.com/doi/10.1111/vru.13358
Smith HE, Schaer TP, Elliott DM, Mauck RL, Mahindroo S, Boyes M, Hilliard R, Meadows K, Fainor M, Orozco BS, and Gullbrand SE. Intervertebral disc degeneration instigates vertebral endplate remodeling and facet joint pathology in a large animal model. European Cells and Materials 2024;47:125-141. https://doi.org/10.22203/eCM.v047a09
Steinmetz JD, Seeher KM, Schiess N, Nichols E, Cao B, Servili C, Cavallera V, Cousin E, Hagins H, Moberg ME, Mehlman ML,… Musaigwa F,… and Dua T. Global, regional, and national burden of disorders affecting the nervous system, 1990–2021: a systematic analysis for the Global Burden of Disease Study 2021. The Lancet Neurology 2024;23:344-381. https://doi.org/10.1016/s1474-4422(24)00038-3
Stewart HL, Gilbert D, Stefanovski D, Garman Z, Albro MB, Bais M, Grinstaff MW, Snyder BD, and Schaer TP. A missed opportunity: A scoping review of the effect of sex and age on osteoarthritis using large animal models. Osteoarthritis Cartilage 2024;32:501-513. https://doi.org/10.1016/j.joca.2024.02.009
Sun X, Dou Z, Shurson GC, and Hu B. Bioprocessing to upcycle agro-industrial and food wastes into high-nutritional value animal feed for sustainable food and agriculture systems. Resources, Conservation and Recycling 2024;201. https://doi.org/10.1016/j.resconrec.2023.107325
Thomas-Hollands A, Hess RS, Weinstein NM, Fromm S, Chappini NA, Marryott K, and Callan MB. Evaluation of post-transfusion RBC alloimmunization in dogs using a gel-column crossmatch with and without anti-canine globulin enhancement. Journal of Veterinary Diagnostic Investigation 2024;36:213-221. https://journals.sagepub.com/doi/10.1177/10406387231222895
Thurston CC, Sertich PL, McDonnell SM, and Parente EJ. Standing hand-assisted laparoscopic ovariohysterectomy to treat chronic pyometra in a mare. J Am Vet Med Assoc 2024;262:1-3. https://doi.org/10.2460/javma.23.11.0646
Tran HH, Watkins A, Oh MJ, Babeer A, Schaer TP, Steager E, and Koo H. Targeting biofilm infections in humans using small scale robotics. Trends Biotechnol 2024;42:479-495. https://doi.org/10.1016/j.tibtech.2023.10.004
Uslu U, Sun L, Castelli S, Finck AV, Assenmacher CA, Young RM, Chen ZJ, and June CH. The STING agonist IMSA101 enhances chimeric antigen receptor T cell function by inducing IL-18 secretion. Nat Commun 2024;15:3933. https://doi.org/10.1038/s41467-024-47692-9
Valle AP, Brown KA, Reilly P, Ciamillo SA, Davidson EJ, Stefanovski D, Stewart HL, and Ortved KF. Effect of video angle on detection of induced front limb lameness in horses. BMC Vet Res 2024;20:172. https://doi.org/10.1186/s12917-024-04032-9
Wang YC, Rassler S, Stefanovski D, Bender J, Deutsch J, Chen T, Cui ZL, and Dou ZX. Evidence of animal productivity outcomes when fed diets including food waste: A systematic review of global primary data. Resources Conservation and Recycling 2024;203. https://doi.org/10.1016/j.resconrec.2024.107411
Wasko UN, Jiang J, Dalton TC, Curiel-Garcia A, Edwards AC, Wang Y, Lee B, Orlen M, Tian S, Stalnecker CA, Drizyte-Miller K, Menard M, Dilly J, Sastra SA, Palermo CF, Hasselluhn MC, Decker-Farrell AR, Chang S, Jiang L, Wei X, Yang YC, Helland C, Courtney H, Gindin Y, Muonio K, Zhao R, Kemp SB, Clendenin C, Sor R, Vostrejs WP, Hibshman PS, Amparo AM, Hennessey C, Rees MG, Ronan MM, Roth JA, Brodbeck J, Tomassoni L, Bakir B, Socci ND, Herring LE, Barker NK, Wang J, Cleary JM, Wolpin BM, Chabot JA, Kluger MD, Manji GA, Tsai KY, Sekulic M, Lagana SM, Califano A, Quintana E, Wang Z, Smith JAM, Holderfield M, Wildes D, Lowe SW, Badgley MA, Aguirre AJ, Vonderheide RH, Stanger BZ, Baslan T, Der CJ, Singh M, and Olive KP. Tumour-selective activity of RAS-GTP inhibition in pancreatic cancer. Nature 2024;629:927-936. https://doi.org/10.1038/s41586-024-07379-z
Whisenant KD, Foucaud M, Mariën T, Levine D, Richardson DW, Stefanovski D, Scherrer NM, Engiles JB, and Ortved K.Dorsal-to-palmar branch neuroanastomosis in horses undergoing palmar digital neurectomy does not reduce neuroma formation or improve outcome. Veterinary Surgery 2024;53:671-683. https://onlinelibrary.wiley.com/doi/10.1111/vsu.14075
Wong HN, Chen TF, Wang PJ, and Holzman LB. ARF6, a component of intercellular bridges, is essential for spermatogenesis in mice. Developmental Biology 2024;508:46-63. https://doi.org/10.1016/j.ydbio.2024.01.007
Wu VV, Swider M, Sumaroka A, Dufour VL, Vance JE, Aleman TS, Aguirre GD, Beltran WA, and Cideciyan AV. Retinal response to light exposure in BEST1-mutant dogs evaluated with ultra-high resolution OCT. Vision Research 2024;218. https://doi.org/10.1016/j.visres.2024.108379
Yang F, Akhtar MN, Zhang D, El-Mayta R, Shin J, Dorsey JF, Zhang L, Xu X, Guo W, Bagley SJ, Fuchs SY, Koumenis C, Lathia JD, Mitchell MJ, Gong Y, and Fan Y. An immunosuppressive vascular niche drives macrophage polarization and immunotherapy resistance in glioblastoma. Sci Adv 2024;10:eadj4678. https://www.science.org/doi/10.1126/sciadv.adj4678
You Y, Missanelli JR, Proctor RM, Haughan J, and Robinson MA. Simultaneous quantification and confirmation of oxycodone and its metabolites in equine urine using ultra-high performance liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2024;1238:124125. https://doi.org/10.1016/j.jchromb.2024.124125
Zavaleta-Martínez A, Barrientos-Morales M, Medina VA, Rodríguez-Andrade A, Cervantes-Acosta P, Hernández-Beltran A, Avendaño-Reyes L, and Domínguez-Mancera B. Evaluation of intrinsic and extrinsic factors affecting pregnancy rate in dual-purpose cows under tropical conditions. Tropical Animal Health and Production 2024;56. https://doi.org/10.1007/s11250-024-04016-9
Zhao G, Gentile ME, Xue LL, Cosgriff CV, Weiner AI, Adams-Tzivelekidis S, Wong JN, Li XY, Kass-Gergi S, Holcomb NP,Basal MC, Stewart KM, Planer JD, Cantu E, Christie JD, Crespo MM, Mitchell MJ, Meyer NJ, and Vaughan AE. Vascular endothelial-derived SPARCL1 exacerbates viral pneumonia through pro-inflammatory macrophage activation. Nature Communications 2024;15. https://doi.org/10.1038/s41467-024-48589-3
Jorge Alvarez
NIH
Mitochondrial influences on blood brain barrier function and neuropsychiatric illness
4/1/2024—10/31/27
$1,898,089
Matthew Atherton
Program in Comparative Animal Biology (PICAB)
PICAB pilot
2/1/24—1/31/25
$30,000
Andres Blanco
NIH
Investigating the role of KAT6A in MLL-rearranged acute myeloid leukemia
9/1/22—8/31/27
$236,549
Michael Hogan
NIH
Defining antiviral T cell responses restricted to the non-classical MHC class Ib molecule, Qa-1
6/1/24—5/31/26
$250,000
Nicola Mason
Sarcoma Gift
Sarcoma
5/1/24—n/a
$50,000
Lisa Murphy
Commonwealth of Pennsylvania
Notifiable avian influenza (NAI) in Pennsylvania
4/1/2024—3/31/2025
$126,223
Jennifer Punt
AAVMC
Advancing veterinary education in Malawi with a focus on research methods and immunology
6/1/2024—5/31/2026
$10,000
Antonella Rotolo
Sarcoma Gift
Sarcoma
5/1/24—n/a
$50,000
Kotaro Sasaki
University of Texas at San Antonio
Advancing brain health research through male germline editing in marmosets
6/1/22—5/31/26
$100,926
Kotaro Sasaki
IRM
IRM translation pilot
1/1/2024—12/31/24
$50,000
Kotaro Sasaki
Quinlivan
Quinlivan gift
1/1/2024—n/a
$50,000
Susan Volk
Drexel University
Roles of type III collagen in the matrix assembly and mechanobiology of cartilage
5/10/2024—4/30/2025
$213,725
More Research News

Food waste: Recycling, not discarding, offers huge environmental benefits (link is external)
Researchers at Penn Vet examine how common food waste recycling methods, including composting, could have a big impact on greenhouse gas emissions and the use of natural resources.

Emmie’s Complex Case Takes Curiosity, Collaboration, and Care
A warmblood named Emmie receives a layered, individualized treatment plan at New Bolton Center for her complex health issues.

A Pennsylvania study of deer and ticks at the University of Pennsylvania School of Veterinary Medicine could shape public health and safety across the Commonwealth
Researchers at the University of Pennsylvania’s School of Veterinary Medicine (Penn Vet), in collaboration with the Pennsylvania Game Commission, are studying the complex interactions between ticks, pathogens, hosts, and their…

