Nearly half of all mammalian genomes originate from ancient retrotransposon integrations. While silenced in nearly all cells, retrotransposon reactivation is a well-known phenomenon in preimplantation embryos and the germline. Reactivation is also observed during epigenetic breakdown that occurs in aging, disease and cancer. Some retrotransposons retained regulatory and structural features that can influence nearby genes. In embryo, these events are transient and some exist less than a cell cycle. A subset of these events splice with nearby protein coding genes, generating embryo and species specific “chimeric transcripts” that form hundreds of novel promoters, exons and polyadenylation sites. Disruptions made in the lab have led to arrested global protein synthesis, cell fate specification errors, stress induced arrest, fertilization failure, embryonic lethality and improper implantation that resembles human pregnancy complications (Cell 2021). This highly collaborative project adapts proteomics, genetics, bioinformatics and CRISPR/Cas9 editing to reveal this overlooked but essential form of retrotransposons-based regulation in development, fertility, and disease.
 As no current cell culture system faithfully represents the preimplantation development, the majority of research at this stage must be done directly in the embryo, and sometimes animal models are necessary. Even with the advent of CRISPR/Cas9 gene editing, generating mouse models is cost prohibitive and largely inaccessible. To circumvent this, I developed a highly efficient electroporation-based editing technique called CRISPR-EZ (CRISPR Electroporation of Zygotes). I showed that CRISPR-EZ is at least 3-4x more efficient than the gold standard of microinjection, is inexpensive, works in all species tested, and can be mastered quickly, making this technology uniquely accessible to many academic labs (JBC 2016, Nature Protocols 2018). While small insertions and deletion strategies have been highly optimized, there is still room to improve in larger insertions, deletion and sophisticated model designs. Current efforts include the use of AAV, CRISPRa/i (activation/interference), humanized models to study conserved developmental regulatory networks.
As no current cell culture system faithfully represents the preimplantation development, the majority of research at this stage must be done directly in the embryo, and sometimes animal models are necessary. Even with the advent of CRISPR/Cas9 gene editing, generating mouse models is cost prohibitive and largely inaccessible. To circumvent this, I developed a highly efficient electroporation-based editing technique called CRISPR-EZ (CRISPR Electroporation of Zygotes). I showed that CRISPR-EZ is at least 3-4x more efficient than the gold standard of microinjection, is inexpensive, works in all species tested, and can be mastered quickly, making this technology uniquely accessible to many academic labs (JBC 2016, Nature Protocols 2018). While small insertions and deletion strategies have been highly optimized, there is still room to improve in larger insertions, deletion and sophisticated model designs. Current efforts include the use of AAV, CRISPRa/i (activation/interference), humanized models to study conserved developmental regulatory networks.
“The Role of Retrotransposon Reactivation in Development and Disease”
1. Study mammalian preimplantation embryos to understand the biological importance of retrotransposon reactivation.
“Junk DNA,” “Jumping Genes,” “Repetitive Elements.” Retrotransposons have been called many things and for decades their expression in early preimplantation was attributed to their selfish nature that takes advantage of an epigenetically defenseless genome. However, we have recently been able to reveal any meaningful function for retrotransposons due to technological advances that reach the single cell level. Single Cell (or Single Embryo) RNA-Seq has revolutionized our ability to accurately track retrotransposon reactivation without the need to pool dozens or hundreds of embryos together, effectively diluting any signal in the noise. We combine publicly available preimplantation datasets across nearly a dozen species together with our own single embryo RNA-Seq data from different genetically altered mouse models in order to use comparative biology to understand the phenomenon of Retrotransposon Reactivation.
Retrotransposon have been associated with various mechanisms, from acting as enhancers or repressors for nearby genes, functioning as topologically associated domains (TADs), or their transcripts contributing to functional Long Non-Coding RNAs (lncRNAs) and splicing with nearby genes in a cell type and stage specific manner (Chimeric Transcripts). Our lab is interested in studying these and other possible mechanisms that operate within mammalian preimplantation embryos, where reactivation is not only highly regulated, but also essential. We then apply this knowledge to other areas of development where retrotransposon reactive (germline, neuronal, immunology, etc) as well as cellular contexts where epigenetic regulation breaks down and can no longer effectively silence retrotransposons (aging, disease and cancer).
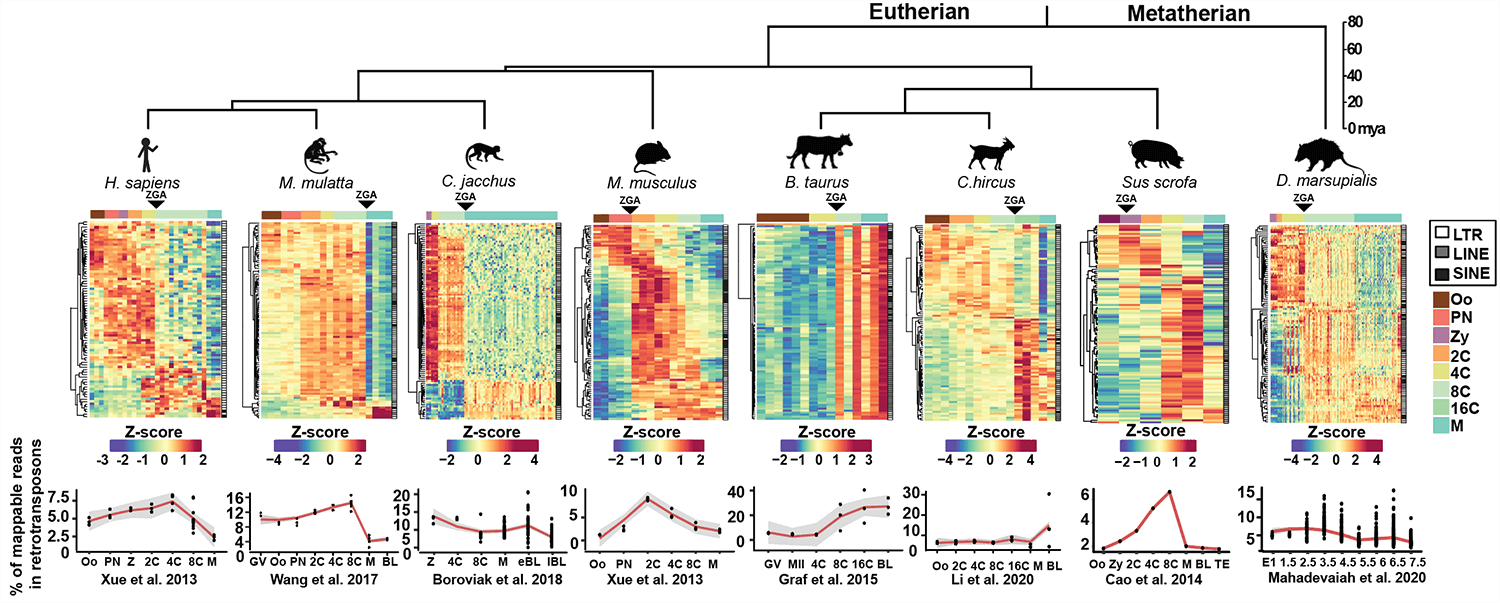 Retrotransposon Reactivation is a conserved phenomenon!
Retrotransposon Reactivation is a conserved phenomenon!
2. Phenotypical analysis and mechanistic understanding of retrotransposon knockout (KO) mouse models.
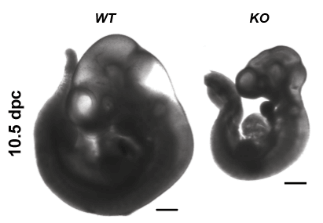 Retrotransposon KO Phenotype figure
Retrotransposon KO Phenotype figure
One of the many fascinating mechanisms that retrotransposons engage in during preimplantation development is the generation of transient and embryo specific splice isoforms between Retrotransposons and protein coding genes called “Chimeric Transcripts”. These are often species specific but about 1/3 appear to be conserved between human and mouse, suggesting potentially important functions. Our previous work (Cell 2021) revealed the first essential retrotransposon in mouse development in the form of MT2B2 acting as a transient promoter driving an embryo specific isoform of Cdk2ap1 (Cdk2ap1MT2B2). This transcript generates an N-terminal truncation (Cdk2ap1ΔN) with potent cell proliferation properties, while the full-length canonical isoform (Cdk2ap1) represses cell proliferation. Deletion of MT2B2, while leaving the Cdkap1 gene intact, reduces cell proliferation and impairs embryo implantation, which leads to both fetal and maternal morbidity. Surprisingly, this isoform is evolutionarily conserved across mammals in sequence and function but is driven by different promoters. In an unusual case of convergent evolution in human and other primates, a completely distinct retrotransposon drives Cdk2ap1ΔN expression!
Cdk2ap1MT2B2 is just one of 5 Retrotransposon deletions made. The other deletions represent promoter, exon, polyadenylation retrotransposon deletions as well as enhancer/repressor mechanisms. Each deletion animal demonstrates unique phenotypical defects ranging from fertilization errors, loss of global translation and severe sensitivity to external stress, morphological defects and loss of pluripotency at the blastocyst stage. Characterization of these mouse models will reveal novel biological pathways and mechanisms that have been rewired by retrotransposon reactivation.
3. Develop Humanized mouse models to study Human specific retrotransposon function.
While the majority of retrotransposon are species specific, their overall expression patterns and impact on nearby genes appears to be conserved. While most of the reactivation events appear to be engaged in species specific mechanisms, a significant proportion (over 30%) are identical between mouse and human, suggesting highly conserved and potentially important developmental functions. Additional animal models are being generated to study these events more closely to reveal pathways and mechanisms that are essential for early development and may contribute to improvements in artificial reproductive technologies (ART) and human health.
Conversely, there are retrotransposon reactivation events that are unique to human embryos, without any evidence in other species. Ectopic expression of these events in mouse embryos to mimic the human condition result changes in embryos, including improvements in viability and cell cycle timing. Additional animal models are being generated in which human retrotransposons are inserted into the orthologous region of the mouse genome to generate humanized mouse models to study their impact and function in early embryos and later in development.
4. Probe embryos, germline, and stem cells to understand retrotransposon regulation.
Another well known (but poorly understood) instance of retrotransposon reactivation occurs during the developmental reprogramming of the male and female germlines. Similar to the early embryo, multiple waves of reactivation occur at distinct developmental stages and can often be used to define timing of oogenesis or spermatogenesis. The reactivation of retrotransposons is a conserved phenomenon observed in the germlines of eukaryotes, spanning worms, flies, and mammals. While failure to properly silence retrotransposons during this brief window of reprogramming often leads to sterility, prevention of reactivation also results in sterility, suggesting that a fine balance between silencing and relaxation of silencing is programmed into proper germline development as well. The mechanisms that allow for precise retrotransposon reactivation in the germline are only just now being elucidated and likely are essential for germline integrity.
In addition to the epigenetic reprogramming that occurs during preimplantation and germline development, a similar phenomenon occurs during the artificial reprogramming of somatic cells, such a fibroblasts, into induced pluripotent stem cells (iPSCs) after introduction of exogenous transcription factors. Recent work has shown that distinct retrotransposons families are induced and are required for somatic reprogramming. Beyond this, ectopic overexpression of certain retrotransposons during this process can create a totipotent-like stem cell state in mouse iPSCs with enhanced differentiation capacity. The mechanisms associated with these processed hold great promise and improvements for regenerative medicine, but the details underlying these mechanisms are almost completely unknown.
5. Explore retrotransposon reactivation consequences outside of the embryo, where epigenetic breakdown occurs, such as aging, disease and cancer.
In addition to the reprogramming that occurs after fertilization, during germline specification, and the artificial reprogramming that occurs to induce pluripotent stem cells (iPSCs), retrotransposons have been observed to reactivate in various developmental contexts. While it is clear that reactivation is intentional and programmed in the developing embryo, the precise nature of reprogramming in other developmental contexts is not clear. Even less obvious is the spontaneous and erratic reactivation events that are associated with the epigenetic breakdown associated with aging, disease and cancer. Under these circumstances, retrotransposon reactivation is prevalent but whether they are drivers or passengers of disease states is up for investigation.
One of our goals is to determine how developmental programs might be “borrowed” during these altered settings. The cell exerts enormous energy on preventing transposon reactivation in somatic cells, but these programs frequently remerge in disease. We would like to help answer the question of whether or not ectopic retrotransposon reactivation contribute to disease prognosis through the re-establishment of dormant developmental programs and if these signatures can be used to more accurately define disease states.
6. Improvements to genome editing technologies (CRISPR-EZ) in mammalian embryos for longer inserts, higher efficiency and lower costs.
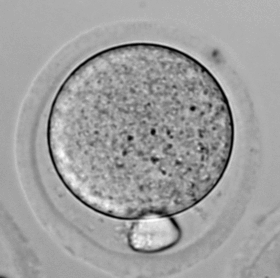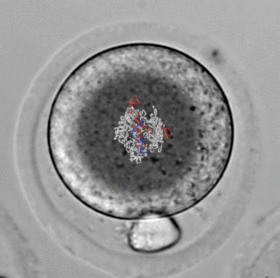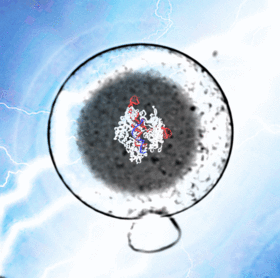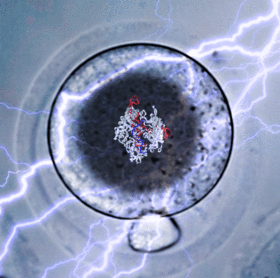 A critical breakthrough in understanding early development is the use of CRISPR/Cas9 systems for the purposes of genome editing, expression modulation, screening, lineage tracing, etc. While these technologies revolutionized modern genetics, they are still difficult techniques to master and inaccessible to many labs. In order to directly manipulate embryos both quickly and cheaply, I developed CRISPR Electroporation of Zygotes (CRISPR-EZ). This electroporation approach bypasses the need for lengthy scheduling of a trained microinjection technician at nearly a fourth of the price.
A critical breakthrough in understanding early development is the use of CRISPR/Cas9 systems for the purposes of genome editing, expression modulation, screening, lineage tracing, etc. While these technologies revolutionized modern genetics, they are still difficult techniques to master and inaccessible to many labs. In order to directly manipulate embryos both quickly and cheaply, I developed CRISPR Electroporation of Zygotes (CRISPR-EZ). This electroporation approach bypasses the need for lengthy scheduling of a trained microinjection technician at nearly a fourth of the price.
Currently, CRISPR-EZ delivers Cas9+guideRNA as a pre-assembled RiboNucleoProtein (RNP) with 100% efficiency for insertion/deletion (in/del) formation. Precise insertions on the order of ~100bp through Homology Directed Repair (HDR) can exceed 60% efficiency. Depending on the size and nature of the desired deletion, this technique can excise between 250bp to 2000bp with an average efficiency of 50%. Larger deletions require significant testing of guide RNA testing and strategies.
Our current efforts are aimed at improving efficiency of deletion sizes and insertion sizes. Reliable insertions greater than 2000bp opens the possibility of tagging proteins, promoter swapping, and generation of conditional alleles. We are testing various viral delivery methods as well as donor oligo chemistries to achieve this industry changing goal. Improvements on this scale would have long lasting and immediate impacts on biomedical research and agriculturally important animal production.
Modzelewski AJ, Shao W, Chen J, Lee A, Qi X, Noon M, Tjokro K, Sales G, Biton A, Anand A, Speed TP, Xuan Z, Wang T, Risso D, He L. A mouse-specific retrotransposon drives a conserved Cdk2ap1 isoform essential for development. Cell. 2021 Oct 28;184(22):5541-5558.e22. doi: 10.1016/j.cell.2021.09.021. Epub 2021 Oct 12. PubMed PMID: 34644528; PubMed Central PMCID: PMC8787082.
Björkgren I, Chung DH, Mendoza S, Gabelev-Khasin L, Petersen NT, Modzelewski A, He L, Lishko PV. Alpha/Beta Hydrolase Domain-Containing Protein 2 Regulates the Rhythm of Follicular Maturation and Estrous Stages of the Female Reproductive Cycle. Front Cell Dev Biol. 2021;9:710864. doi: 10.3389/fcell.2021.710864. eCollection 2021. PubMed PMID: 34568325; PubMed Central PMCID: PMC8455887.
Rosàs-Canyelles E*, Modzelewski AJ*, Gomez Martinez AE, Geldert A, Gopal A, He L, Herr AE. Multimodal detection of protein isoforms and nucleic acids from low starting cell numbers. Lab Chip. 2021 Jun 15;21(12):2427-2436. doi: 10.1039/d1lc00073j. PubMed PMID: 33978041; PubMed Central PMCID: PMC8206029.
Modzelewski A, Shao W, Chen J, Lee A, Qi X, Noon M, Tjokro K, Sales G, Biton A, Speed T, Xuan Z, Wang T, Risso D, He L. A species-specific retrotransposon drives a conserved <i>Cdk2ap1</i> isoform essential for preimplantation development. [preprint]. 2021 March. doi: 10.1101/2021.03.24.436683.
Rosàs-Canyelles E*, Modzelewski AJ*, Geldert A, He L, Herr AE. Multimodal detection of protein isoforms and nucleic acids from mouse pre-implantation embryos. Nat Protoc. 2021 Feb;16(2):1062-1088. doi: 10.1038/s41596-020-00449-2. Epub 2021 Jan 15. PubMed PMID: 33452502; PubMed Central PMCID: PMC7954398.
Tsitsiklis A, Bangs DJ, Lutes LK, Chan SW, Geiger KM, Modzelewski AJ, Labarta-Bajo L, Wang Y, Zuniga EI, Dai S, Robey EA. An Unusual MHC Molecule Generates Protective CD8+ T Cell Responses to Chronic Infection. Front Immunol. 2020;11:1464. doi: 10.3389/fimmu.2020.01464. eCollection 2020. PubMed PMID: 32733483; PubMed Central PMCID: PMC7360836.
Modzelewski AJ, Chen S, Willis BJ, Lloyd KCK, Wood JA, He L. Efficient mouse genome engineering by CRISPR-EZ technology. Nat Protoc. 2018 Jun;13(6):1253-1274. doi: 10.1038/nprot.2018.012. Epub 2018 May 10. PubMed PMID: 29748649; PubMed Central PMCID: PMC6296855.
Hilz S, Fogarty EA, Modzelewski AJ, Cohen PE, Grimson A. Transcriptome profiling of the developing male germ line identifies the miR-29 family as a global regulator during meiosis. RNA Biol. 2017 Feb;14(2):219-235. doi: 10.1080/15476286.2016.1270002. Epub 2016 Dec 16. PubMed PMID: 27981880; PubMed Central PMCID: PMC5324742.
Hilz S*, Modzelewski AJ*, Cohen PE, Grimson A. The roles of microRNAs and siRNAs in mammalian spermatogenesis. Development. 2016 Sep 1;143(17):3061-73. doi: 10.1242/dev.136721. Review. PubMed PMID: 27578177; PubMed Central PMCID: PMC5047671.
Chen S, Lee B, Lee AY, Modzelewski AJ‡, He L‡. Highly Efficient Mouse Genome Editing by CRISPR Ribonucleoprotein Electroporation of Zygotes. J Biol Chem. 2016 Jul 8;291(28):14457-67. doi: 10.1074/jbc.M116.733154. Epub 2016 May 5. PubMed PMID: 27151215; PubMed Central PMCID: PMC4938170.
Sun X, Brieño-Enríquez MA, Cornelius A, Modzelewski AJ, Maley TT, Campbell-Peterson KM, Holloway JK, Cohen PE. FancJ (Brip1) loss-of-function allele results in spermatogonial cell depletion during embryogenesis and altered processing of crossover sites during meiotic prophase I in mice. Chromosoma. 2016 Jun;125(2):237-52. doi: 10.1007/s00412-015-0549-2. Epub 2015 Oct 21. PubMed PMID: 26490168; PubMed Central PMCID: PMC5415080.
Modzelewski AJ, Hilz S, Crate EA, Schweidenback CT, Fogarty EA, Grenier JK, Freire R, Cohen PE, Grimson A. Dgcr8 and Dicer are essential for sex chromosome integrity during meiosis in males. J Cell Sci. 2015 Jun 15;128(12):2314-27. doi: 10.1242/jcs.167148. Epub 2015 May 1. PubMed PMID: 25934699; PubMed Central PMCID: PMC4487015.
Zhang J, Bonasio R, Strino F, Kluger Y, Holloway JK, Modzelewski AJ, Cohen PE, Reinberg D. SFMBT1 functions with LSD1 to regulate expression of canonical histone genes and chromatin-related factors. Genes Dev. 2013 Apr 1;27(7):749-66. doi: 10.1101/gad.210963.112. PubMed PMID: 23592795; PubMed Central PMCID: PMC3639416.
Lyndaker AM, Modzelewski AJ, Welsh IC. Reproductive and developmental genomics retreat at Cornell University, 2012. Mol Reprod Dev. 2012 Sep;79(9):588-91. doi: 10.1002/mrd.22081. Epub 2012 Aug 29. Review. PubMed PMID: 22933246; PubMed Central PMCID: PMC4104196.
Modzelewski AJ, Holmes RJ, Hilz S, Grimson A, Cohen PE. AGO4 regulates entry into meiosis and influences silencing of sex chromosomes in the male mouse germline. Dev Cell. 2012 Aug 14;23(2):251-64. doi: 10.1016/j.devcel.2012.07.003. Epub 2012 Aug 2. PubMed PMID: 22863743; PubMed Central PMCID: PMC3470808.
Holloway JK, Mohan S, Balmus G, Sun X, Modzelewski A, Borst PL, Freire R, Weiss RS, Cohen PE. Mammalian BTBD12 (SLX4) protects against genomic instability during mammalian spermatogenesis. PLoS Genet. 2011 Jun;7(6):e1002094. doi: 10.1371/journal.pgen.1002094. Epub 2011 Jun 2. PubMed PMID: 21655083; PubMed Central PMCID: PMC3107204.
Meet the Modzelewski Lab Team
 |
Andrew Modzelewski, PhD, Principal Investigator
Dr. Modzelewski is a developmental biologist with an interest in early mammalian development and male and female fertility. He is particularly interested in understanding how retrotransposon function in these important cellular contexts. He is also passionate about developing tool to study preimplantation embryos and always has a fun side project to keep these technologies innovative.
He studied Biochemistry and Biophysics at the Pennsylvania State University (PSU) where he got his BS in Biochemistry and Molecular Biology and minored in chemistry and philosophy. He developed a deep interest in reproduction at Cornell University (PhD) and moved into preimplantation development as a postdoctoral fellow at the University of California, Berkeley. He moved to Philadelphia in 2022 to start his own lab to tie all these previous experiences together.
Dr. Modzelewski believes that retrotransposon reactivation is essential for proper development and is in fact a part of “Normal Biology”. By studying retrotransposons in the embryo, where the phenomenon of retrotransposon reactivation is intentional and essential, we can learn more about how the unintentional retrotransposon reactivations observed in aging, infertility and cancer contribute to malignancies and where this can be mitigated or even reserved.
|
 |
Chantal Diallo
|